
Articles by rOpenSci - open tools for open science
One of the first things I took on when I started at the Cornell Lab of Ornithology was creating the auk R package for accessing eBird data. The entire eBird dataset can be downloaded as a massive text file, called the eBird Basic Dataset (EBD), and auk pulls out manageable ... [Read more...]
rOpenSci Dev Guide 0.4.0: Updates
rOpenSci Software Peer Review’s guidance has been compiled in an online book for more than one year now. We’ve just released its fourth version.
To find out what’s new in our dev guide 0.4.0, you can read the changelog,
or this blog post for more digested information.
Note ... [Read more...]
10 Things We Learned in Creating the Blog Guide with bookdown
After soliciting, reviewing, and publishing over 100 blog posts and tech notes by rOpenSci community members, we have created the rOpenSci Blog Guide for Authors and Editors to address many frequently asked questions and frequently given suggestions.
Technically, we structured the content as a bookdown gitbook. It was Stef’s first ... [Read more...]
parzer: Parse Messy Geographic Coordinates
parzer is a new package for
handling messy geographic coordinates. The first version is now on CRAN,
with binaries coming soon hopefully (see note about installation below).
The package recently completed rOpenSci
review.
parzer motivation
The idea for this package started with a tweet from Noam
Ross
(https://twitter.com/... [Read more...]
Supercharge your GitHub Actions Experience with tic
“Continuous Integration” (CI) has become a standard for proper software development.
Checking the integrity of software after changes have been made is essential to ensure its proper functionality.
Also, CI helps catch problems introduced by dependencies early when executed on a regular basis (usually done via scheduled CRON runs).
Multiple ... [Read more...]
Community Call – Maintaining an R Package
For this Community Call, we’re trying something different. We’ll start with a short talk by Julia Silge, then spend most of the time on Q & A with four panelists - Elin Waring, Erin Grand, Leonardo Collado-Torres, and Scott Chamberlain - moderated by Julia.
Our panelists bring a wide ...

opentripplanner: Fast and Easy Multimodal Trip Planning in R with OpenTripPlanner
With services like Google Maps, finding the fastest route from A to B has become quick, cheap, and easy. Not just for driving but walking, cycling and public transport too. But in the field of transport studies, we often want not only a single route, but thousands or millions of ... [Read more...]
rOpenSci’s Leadership in #rstats Culture
At their closing keynote at the 2020 RStudio Conference, Hilary Parker and Roger Peng mentioned that they hatched the idea for their excellent Not So Standard Deviations podcast following their reunion at the 2015 rOpenSci unconf, (“runconf15”). That statement went straight to my heart because it pin-pointed how I had been feeling ...
2 Months in 2 Minutes – rOpenSci News, February 2020
rOpenSci HQ
On behalf of rOpenSci, thank you to everyone who has contributed their creativity, curiosity, smarts, and time in the last year. Read our Thank You, 2019.
Software Peer Review
3 community-contributed packages passed... [Read more...]
taxadb: A High-Performance Local Taxonomic Database Interface
Dealing with taxonomic inconsistencies within and across datasets is a fundamental challenge of ecology and evolutionary biology. Accounting for species synonyms, taxa splitting and unification is especially important as aggregation of data across time and different data sources becomes increasingly common. One potentially powerful approach for addressing these issues is ... [Read more...]
The Fun of Building Things and the Challenge of Learning – the rOpenSci OzUnconf 2019
It was the best of times, it was the worst of times.
Dickens might have meant it figuratively, but in the case of the rOpenSci OzUnconf 2019, we mean it literally. Set to the backdrop of a national emergency that is still ongoing from 11-13 December, our participants came from across ... [Read more...]
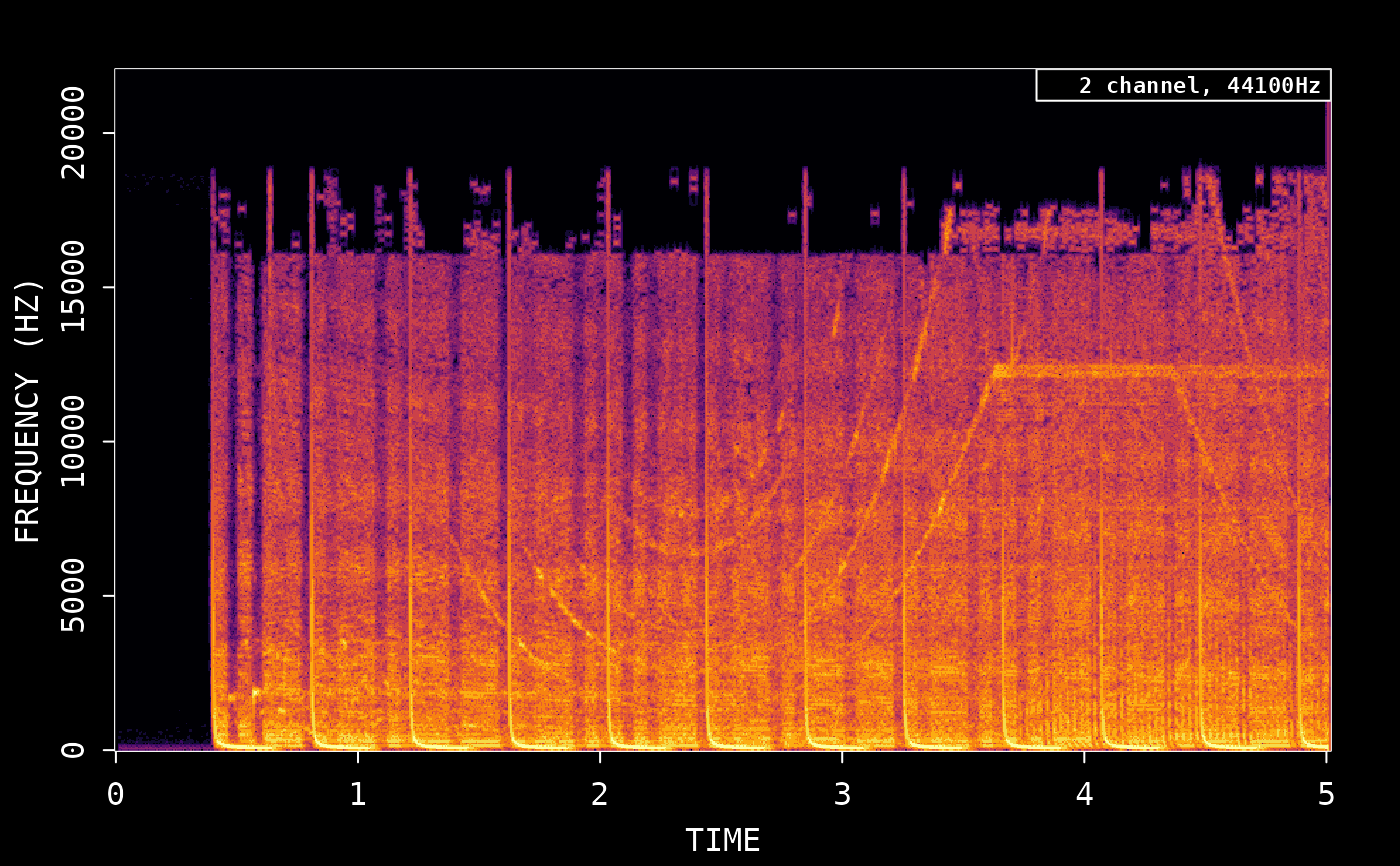
Working with audio in R using av
The latest version of the rOpenSci av package includes some useful new tools for working with audio data. We have added functions for reading, cutting, converting, transforming, and plotting audio data in any popular audio / video format (mp3, mkv, aac, etc).
The functionality can either be used by itself, or ...
Call BEAST2 for Bayesian evolutionary analysis from R
babette 1 is a package to work with BEAST2 2,
a software platform for Bayesian evolutionary analysis from R.
babette is a spin-off of my own academic research.
As a PhD I work on models of diversification: mathematical descriptions
of how species form new species. Instead of working on a species’
individuals, ... [Read more...]
rOpenSci Code of Conduct Annual Review
One year ago, we released our new Code of Conduct. At that time, the Code of Conduct Committee (authors of this post) agreed to do an annual review of the text, reporting form, and our internal guidelines.
We have made one change to the Code of Conduct text. Because some ... [Read more...]
rOpenSci 2019 Code of Conduct Transparency Report
In January 2019, we announced the release of rOpenSci’s Code of Conduct version 2.0. This includes a named Committee, greater detail about unacceptable behaviors, instructions on how to make a report, and information on how reports are handled. We are committed to transparency with our community while upholding of victims and ... [Read more...]
Thank You, 2019
We mean it.
On behalf of rOpenSci, thank you to everyone who has contributed their creativity, curiosity, smarts, and time in the last year. We are fortunate to have paid staff who work to build technical and social infrastructure to... [Read more...]
Want to Intern with rOpenSci’s Community Manager?
Want to get some hands-on insights into running an open source community? Here’s an opportunity to work with me, rOpenSci’s Community Manager, on some non-code community-related work. I am looking for someone to work 1 day a week ... [Read more...]
2 Months in 2 Minutes – rOpenSci News, December 2019
rOpenSci HQ
rOpenSci Announces a New $896k Award From The Gordon and Betty Moore Foundation to Improve the Scientific Package Ecosystem for R.
We’re excited to announce a new member of our team! Introducing Mark Padgham, rOpenSci’s new Software Research Scientist
NumFOCUS recognizes Melina Vidoni and Will Landau ... [Read more...]
We cleaned our website URLs with R!
Last year we reported on the joy of using commonmark and xml2 to parse
Markdown content, like the source of this website built with
Hugo, in particular to extract
links,
at the time merely to count them. How about we go a bit further and use
the same approach to ... [Read more...]
HTTP testing in R: overview of tools and new features
Testing is a crucial component to any software package. Testing makes sure
that your code does what you expect it to do; and importantly, makes it safer to make
changes moving forward because a good test suite will tell you if a change has broken
existing functionality. Our recent community ... [Read more...]
Copyright © 2022 | MH Corporate basic by MH Themes