A modular Rmarkdown workbook in action I’m now using a…
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
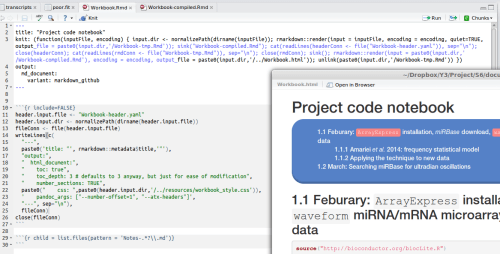


A modular Rmarkdown workbook in action
I’m now using a properly modular Rmarkdown workbook, and really quite chuffed with it! I’ve seen discussion of electronic lab notebooks for bioinformatics with only mention of org-mode for emacs users, IPython (now the Jupyter project) and the likes of Evernote/Word etc., so I’m sharing the basic workings of mine.
Here’s its custom knit function, of the kind I wrote about the other day:
The workflow :
When the knit button is clicked (or
Ctrl+Shift+K), the document renders itself to markdown, and the notes specified as ‘children’ (in the chunk at the bottom of the top picture) are stuck onto the end. These individual notes are written from Rmarkdown themselves with YAML headers:This header just specifies to knit quietly (i.e. not preview the resulting markdown) and pass on an instruction to pandoc to use Atx-style headers (the ones with
#symbols, needed for proper table of contents numbering down the line). The R code that follows it prints the title as this sort of [second-level] header.…during that knitting process the big code chunk in the top picture writes a new header configuration file
Workbook-header.yaml(essential for a proper table of contents)sinkallows this header to be concatenated with the compiled workbook,Workbook-compiled-tmp.Rmd- Lastly the HTML workbook is written and previewed, and the temporary (header-less) compilation file is removed with
unlink. I chose to keep the full compiledWorkbook-compiled.Rmdfile for the time being. Custom CSS styling for the Table of Contents is below and here, along with all the other code snippets in this post.
I’ll add any further changes to my development notes Wiki. Thanks to Vincent Nijs for the
childcode chunk tip as an issue in the repo there 🙂 More comments/suggestions through that route welcome
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
