MCMC chain analysis and convergence diagnostics with coda in R
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
Last week, I gave a seminar about MCMC chain analysis and convergence diagnostics with coda in R, and I thought a summary would make a nice post.
As a prerequisite, we will use a few lines of code, very similar to a previous post on MCMC sampling. In the code, we create some test data in which a dependent variable y is in a more or less linear relationship with an independent variable x; define likelihood and priors for a linear model to be fit to the data; and implement a simple Metropolis-Hastings MCMC to sample from the posterior distribution of this model. It’s actually not important for the things that follow that you understand in detail what’s going on here, but for those who do not feel at ease about this, have a look at my previous post.
library(coda)
trueA <- 5
trueB <- 0
trueSd <- 10
sampleSize <- 31
x <- (-(sampleSize-1)/2):((sampleSize-1)/2)
y <- trueA * x + trueB + rnorm(n=sampleSize,mean=0,sd=trueSd)
likelihood <- function(param){
a = param[1]
b = param[2]
sd = param[3]
pred = a*x + b
singlelikelihoods = dnorm(y, mean = pred, sd = sd, log = T)
sumll = sum(singlelikelihoods)
return(sumll)
}
prior <- function(param){
a = param[1]
b = param[2]
sd = param[3]
aprior = dunif(a, min=0, max=10, log = T)
bprior = dnorm(b, sd = 5, log = T)
sdprior = dunif(sd, min=0, max=30, log = T)
return(aprior+bprior+sdprior)
}
proposalfunction <- function(param){
return(rnorm(3,mean = param, sd= c(0.1,0.5,0.3)))
}
run_metropolis_MCMC <- function(startvalue, iterations){
chain = array(dim = c(iterations+1,3))
chain[1,] = startvalue
for (i in 1:iterations){
proposal = proposalfunction(chain[i,])
probab = exp(likelihood(proposal)+ prior(proposal) - likelihood(chain[i,])- prior(chain[i,]))
if (runif(1) < probab){
chain[i+1,] = proposal
}else{
chain[i+1,] = chain[i,]
}
}
return(mcmc(chain))
}
So, let’s run the MCMC:
chain = run_metropolis_MCMC(startvalue, 10000)
The run_metropolis_MCMC() function basically returns a posterior sample created by the MCMC algorithm as an array with one column for each parameter and as many rows as there are steps in the MCMC. This it is a little hard to see because the function return is already formatted as a coda mcmc object (the line “return(mcmc(chain))” at the end of the function transforms the chain into a coda object). Yet, you still see the structure either by str(chain), or by transforming the coda object back to a normal data-frame by data.frame(chain).
Some simple summaries of the chain
The advantage of having a coda object is that a lot of things that we typically want to do with the chain are already implemented, so for example we can simply summary() and plot() the outputs
summary(chain) plot(chain)
which gives some useful information on the console and a plot that should look roughly like this:
Figure: Results of the plot() function of a coda object
I think the summary is self-explanatory (otherwise check out the help), but maybe a few words on the results of the plot() function: each row corresponds to one parameter, so there a are two plots for each parameter. The left plot is called a trace plot – it shows the values the parameter took during the runtime of the chain. The right plot is usually called a marginal density plot. Basically, it is the (smoothened) histogram of the values in the trace-plot, i.e. the distribution of the values of the parameter in the chain.
Marginal densities hide correlations
Marginal densities are an average over the values a parameter takes with all other parameters “marginalized”, i.e. other parameters having any values according to their posterior probabilities. Often, marginal densities are treated as the main output of a Bayesian analysis (e.g. by reporting their mean and sd of), but I strongly advice against this practice without further analysis. The reason is that marginal densities “hide” correlations between parameters, and if there are correlations, parameter uncertainties appear to be much greater in the marginals that they actually are. To check for pairwise correlations is quite easy – just use pairs on the MCMC chain:
pairs(data.frame(chain))
or use this code snippet which produces the “nicer” pair correlation plot that you can see below.
In our case, there should be no large correlations because I set up the example in that way, but we can easily achieve a correlation between slope and intercept by “unbalancing” our data, that is, having x-values that are not centered around 0. To see this, replace in the first large code fragment the creation of the test data by this line which creates non-centered x-values
x <- (-(sampleSize-1)/2):((sampleSize-1)/2) + 20
Running the MCMC again and checking for correlations should give you a quite different picture. I show an example below, plotted with a bit fancier plot function than pairs:
Figure: Marginal densities (diagonal), pairwise densities (lower panes) and correlation coefficien (upper panels) for the fit with unbalanced x-values
You can see the strong correlation between the first and the second parameter (slope and intercept), and you can also see that your marginal uncertainty for each parameter (on the diagonal, or in your plot() function) has increased. However, this is not because the fit is fundamentally more uncertain – the Bayesian analysis doesn’t care if you shift the x-values by 20 to the right. Unlike some other statistical techniques, the fundamental fit in terms of the chain has no problems with the correlations. However, it is problematic now to summarize the results of such an analysis e.g. in terms of marginal values. For example, it doesn’t make sense any more to say that the slope has a value of x +/- sd because this misses that point that for any given parameter of the intercept, the uncertainty of the slope is much smaller. For that reason, one should always check the correlations, and if possible, one should try to avoid correlations between parameters because this makes the analysis easier.
Note that we only checked for pairwise correlations here, there may still be higher order interactions that don’t show up in an analysis like that, so you may still be missing something. For that reason, the advice is to summarize the chain only when really necessary, otherwise things like the prior predictive distribution etc. should always be created by sampling directly from the chain.
Convergence diagnostics
Now, to the convergence: an MCMC creates a sample from the posterior distribution, and we usually want to know whether this sample is sufficiently close to the posterior to be used for analysis. There are several standard ways to check this, but I recommend the Gelman-Rubin diagnostic (check the coda help for other options that are implemented). Basically, Gelman-Rubin measures whether there is a significant difference between the variance within several chains and the variance between several chains by a value that is called “scale reduction factors”. To do this, we obviously need a second chain, and then simply run the commands:
chain2 = run_metropolis_MCMC(startvalue, 10000) combinedchains = mcmc.list(chain, chain2) plot(combinedchains) gelman.diag(combinedchains) gelman.plot(combinedchains)
The plot should look something like this
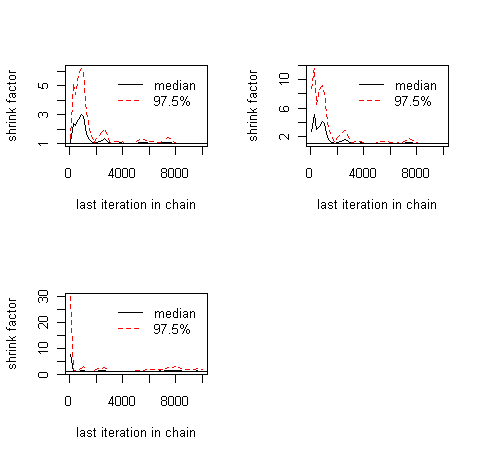
Figure: Results of the gelman.plot() function
The gelman.diag gives you the scale reduction factors for each parameter. A factor of 1 means that between variance and within chain variance are equal, larger values mean that there is still a notable difference between chains. Often, it is said that everything below 1.1 or so is OK, but note that this is more a rule of thumb. The gelman,plot shows you the development of the scale-reduction over time (chain steps), which is useful to see whether a low chain reduction is also stable (sometimes, the factors go down and then up again, as you will see). Also, note that for any real analysis, you have to make sure to discard any bias that arises from the starting point of your chain (burn-in), typical values here are a few 1000-10000 steps. The gelman plot is also a nice tool to see roughly where this point is, that is, from which point on the chains seem roughly converged.
Improving convergence / mixing
So, what to do if there is no convergence yet? Of course, you can always run the MCMC longer, but the other option is to make it converge faster … the word that is used here is “mixing”, which basically means how well the algorithm jumps around in the parameter space … the mixing is affected by the choice of your proposal function. Two things can happen:
- Your proposal function is narrow compared to the distribution we sample from – high acceptance rate, but we don’t get anywhere, bad mixing
- Your proposal function is too wide compared to the distribution we sample from – low acceptance rate, most of the time we stay where we are
Figure: problems in the proposal function and how they show up in trace-plots
As displayed in the figure, these problems can be seen in the trace plots. Theoretical considerations show that an acceptance rate of 20-30% is optimal for typical target distributions, but this is in practice not so helpful because you can still have very bad mixing although being at this level by having the proposal of one parameter too narrow and the proposal of another parameter too wide. Better to look at the trace-plots of the individual parameters. Again, correlations are a problem, so if you have strong correlations in parameter space, you can get bad mixing. Using multivariate proposals that are adjusted to the correlation structure can help, but in general it is better to avoid correlations if possible. The good news at last: most of the more “professional” MCMC sampling software such as Jags or WinBugs will do these things automatically for you.
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.