
Articles by R on The broken bridge between biologists and statisticians
Pairwise comparisons are one of the most debated topic in agricultural research: they are very often used and, sometimes, abused, in literature. I have nothing against the appropriate use of this very useful technique and, for those who are interest... [Read more...]
Regression analyses with common checks in pesticide research
In pesticide research or, in general, agriculture research, we very commonly encounter experiments with, e.g., several herbicides tested at different doses and in different conditions. For these experiments, the untreated control is always added and... [Read more...]
Factorial designs with check in pesticide research
In pesticide research or, in general, agriculture research, we very commonly encouter experiments with two/three crossed factors and some other treatment that is not included in the factorial structure. For example, let’s consider an experiment with... [Read more...]
Repeated measures and subsampling with perennial crops
In a recent post, I have talked about repeated measures, for a case where measurements were taken repeatedly in the same plots across years see here. Previously, in another post, I had talked about subsampling, for a case where several random sample... [Read more...]
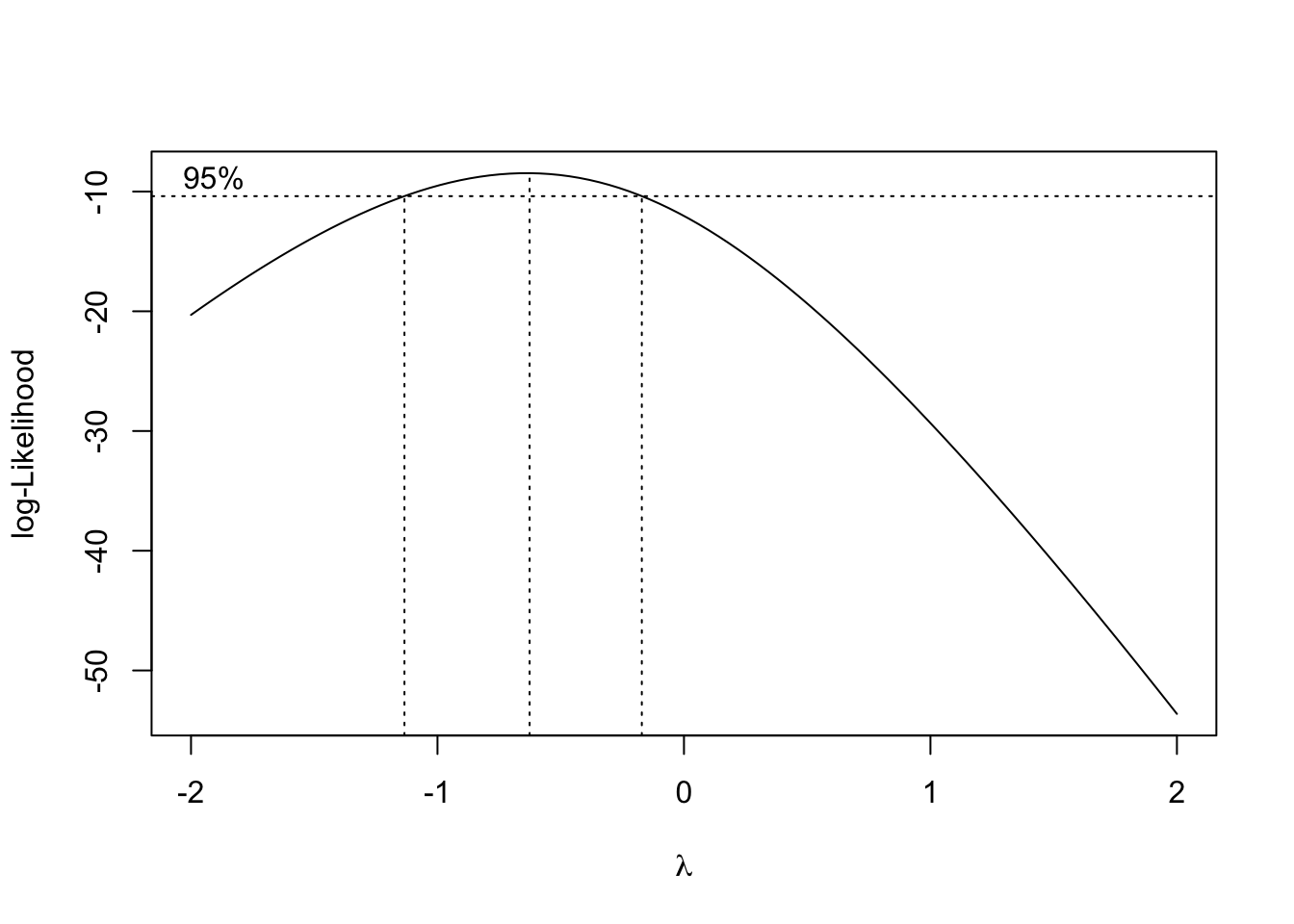
Back-transformations with emmeans()
I am one of those old guys who still uses the stabilising transformations, when the data do not conform to the basic assumptions for ANOVA. Indeed, apart from counts and proportions, where GLMs can be very useful, I have not yet found a simple way t...
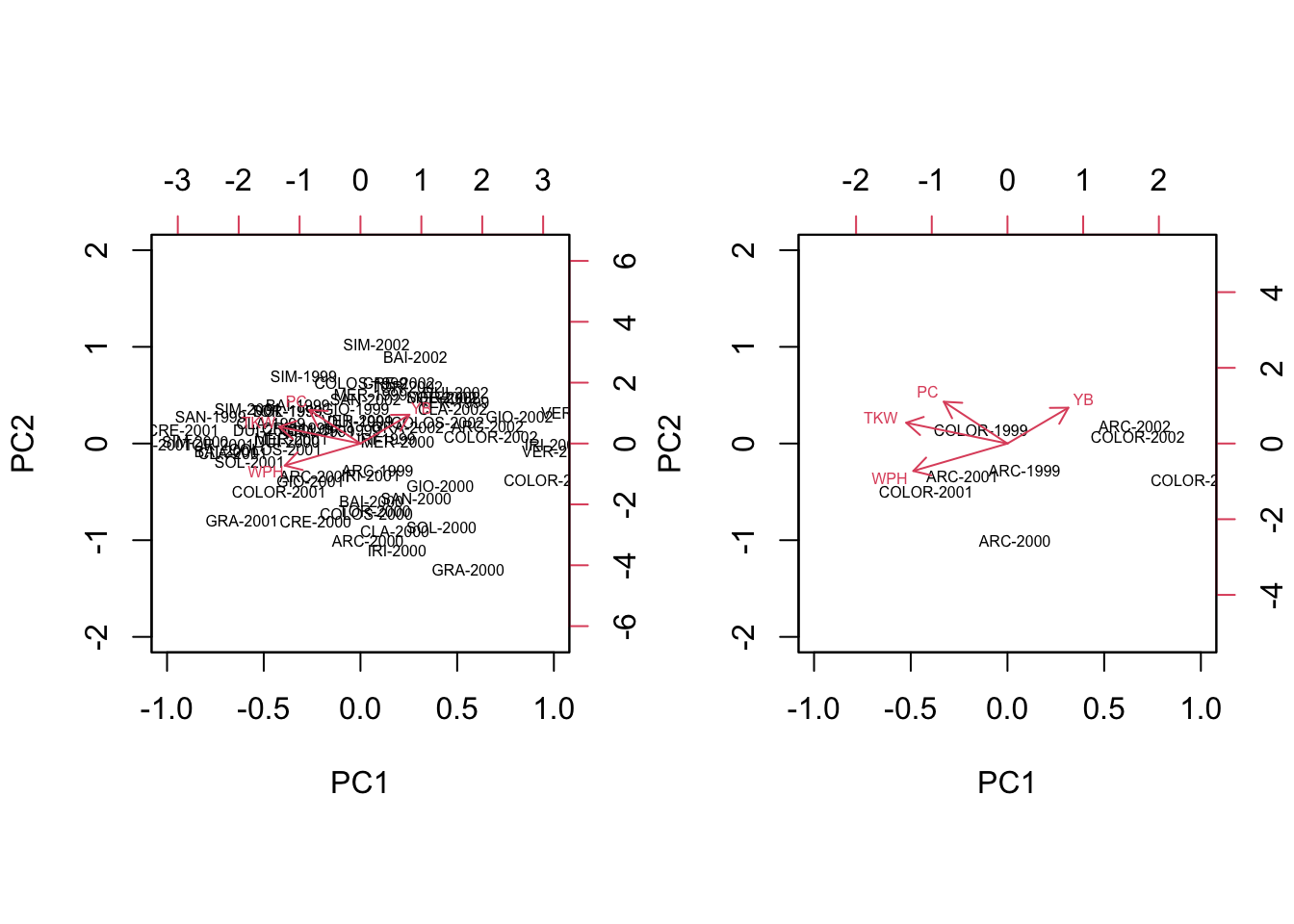
Designed experiments with replicates: Principal components or Canonical Variates?
A few days ago, a colleague of mine wanted to hear my opinion about what multivariate method would be the best for a randomised field experiment with replicates. We had a nice discussion and I thought that such a case-study might be generally intere...
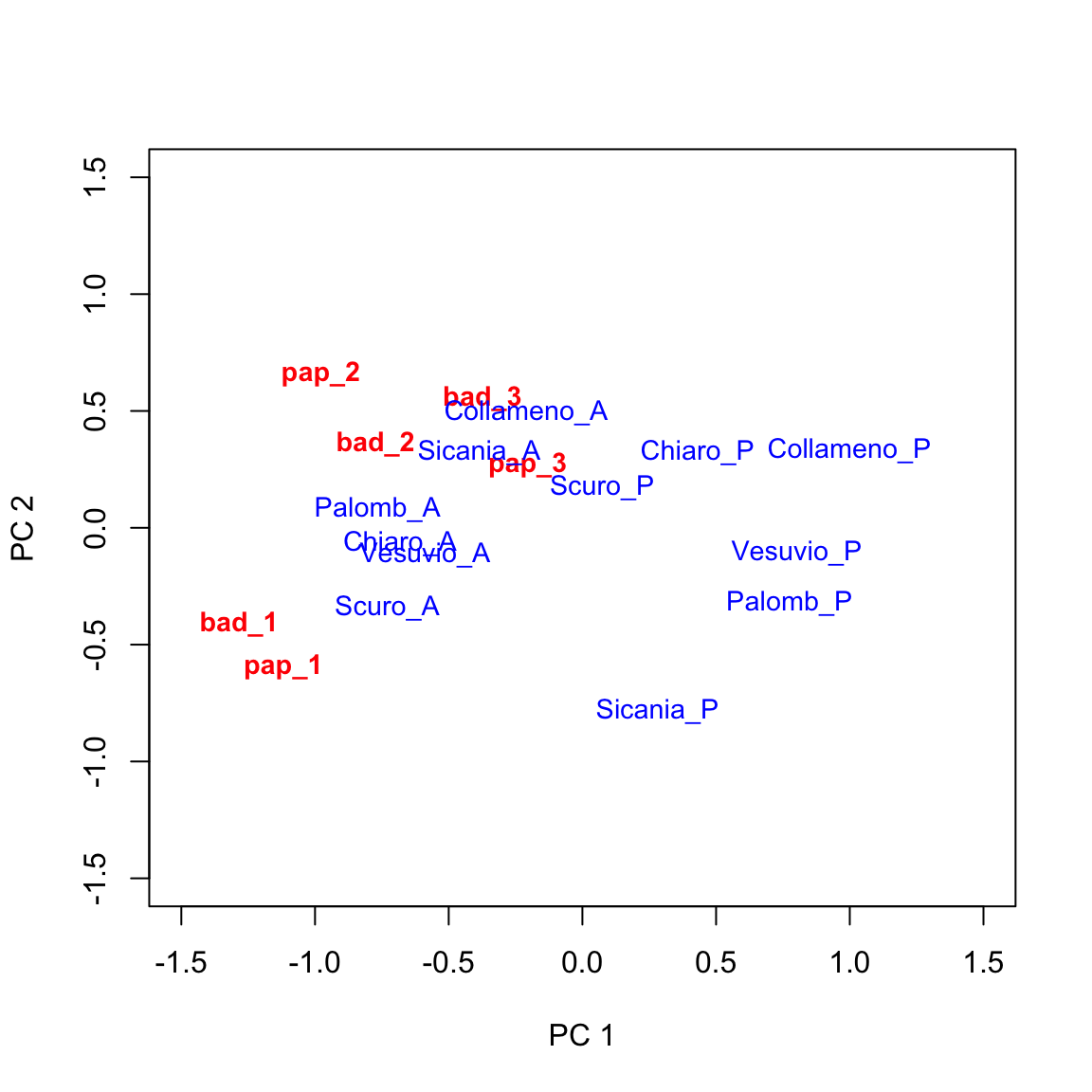
GGE analyses for multi-environment studies
In a recent post we have seen that we can use Principal Component Analyses (PCA) to elucidate the ‘genotype by environment’ relationship (see this post). Whenever the starting point for PCA is the doubly-centered (centered by rows and columns) matri...
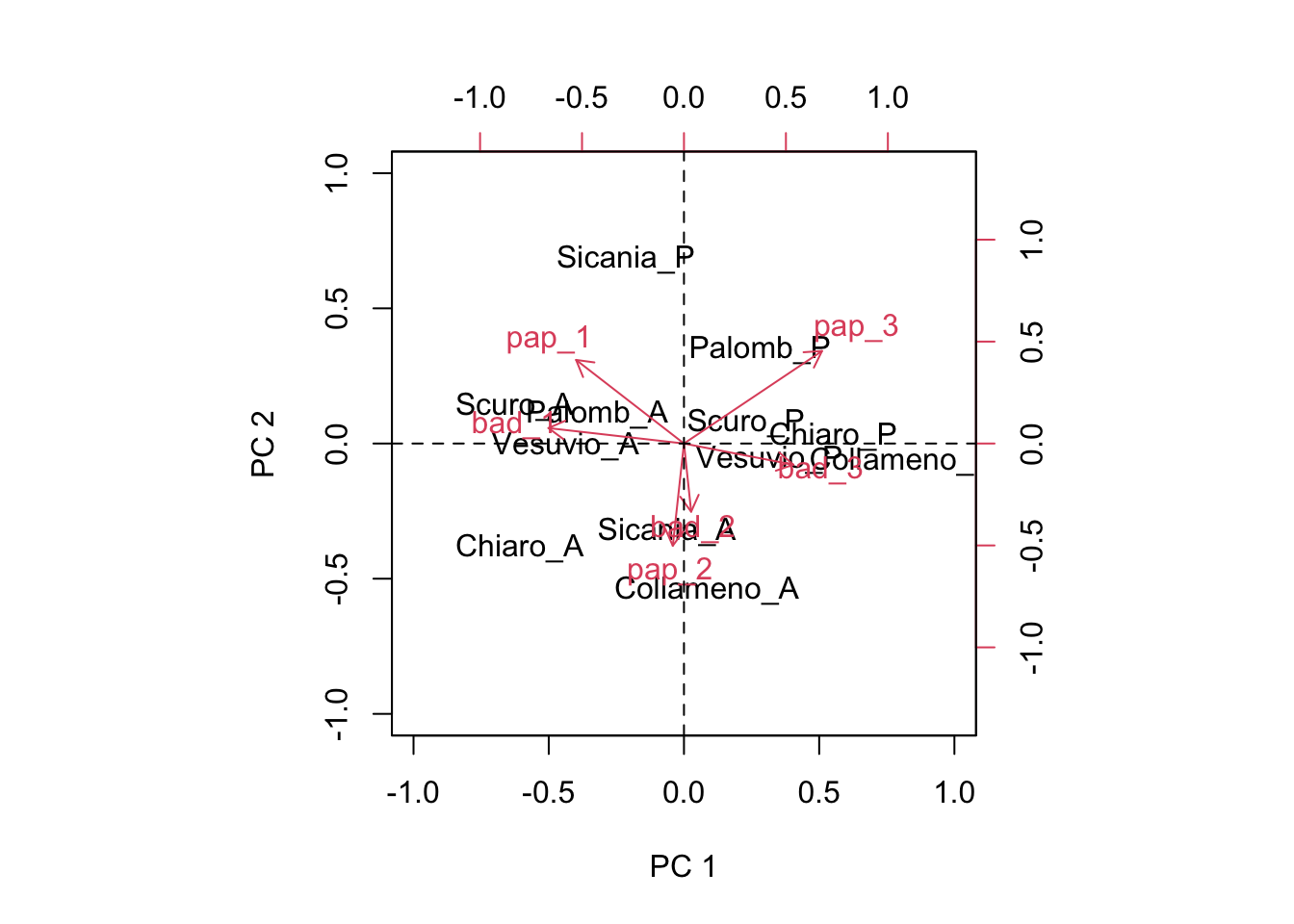
AMMI analyses for multi-environment studies
Again into a subject that is rather important for most agronomists, i.e. the selection of crop varieties. All farmers are perfectly aware that crop performances are affected both by the genotype and by the environment. These two effects are not pure...
Repeated measures with perennial crops
In this post, I want to discuss a concept that is often mistaken by some of my collegues. With all crops, we are used to repeating experiments across years to obtain multi-year data; the structure of the resulting dataset is always the same and it i... [Read more...]
Subsampling in field experiments
Subsampling is very common in field experiments in agriculture. It happens when we collect several random samples from each plot and we submit them to some sort of measurement process. Some examples? Let’s look at the following (very incomplete) lis... [Read more...]
Fitting threshold models to seed germination data
In previous posts we have shown that we can use time-to-event curves to describe the germination pattern of a seed population (see here). We have also shown that these curves can be modified to include the effects of external/internal factors/covariates, such as the genotype, the species, the humidity ...
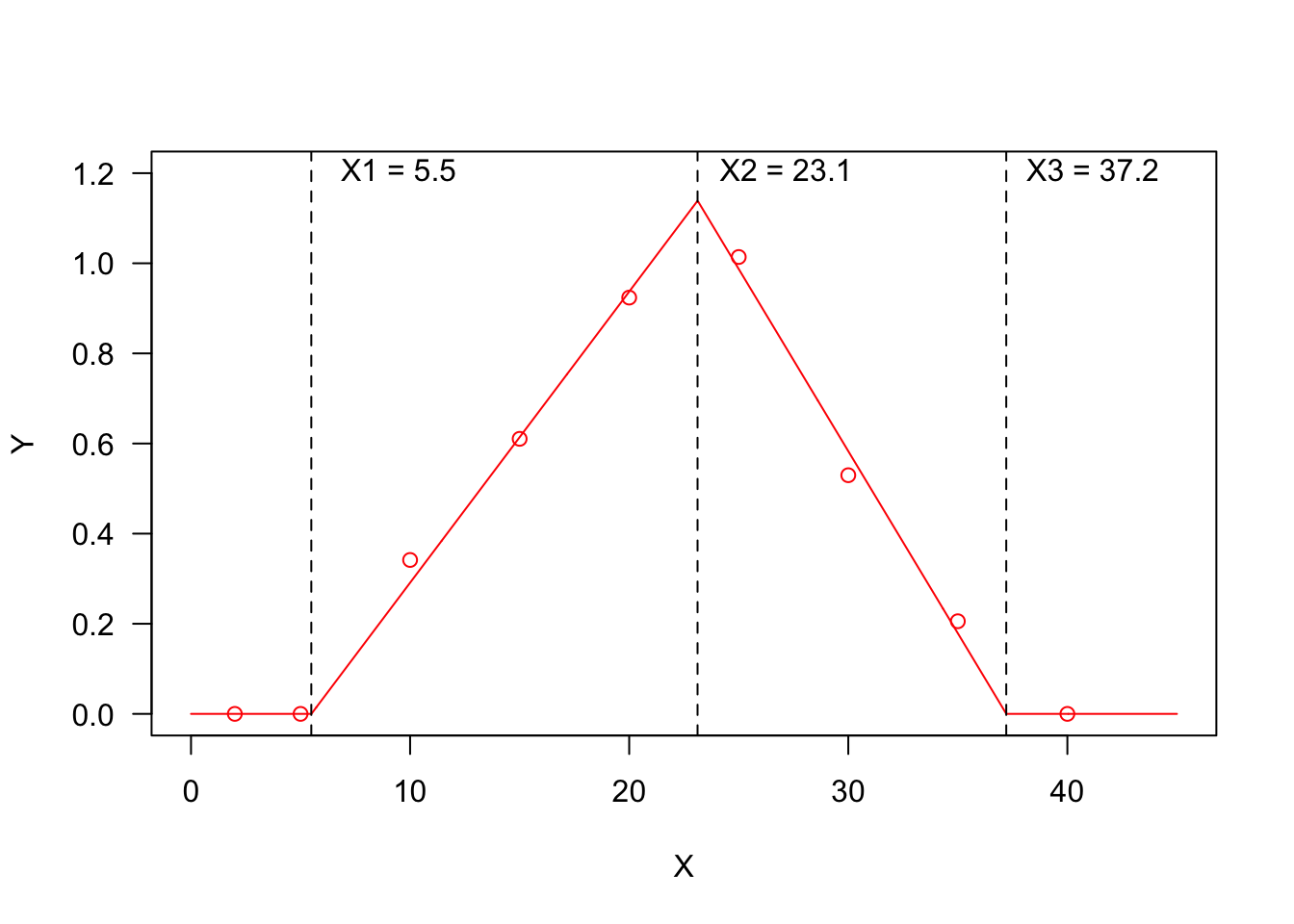
Fitting thermal-time-models to seed germination data
This is a follow-up post. If you are interested in other posts of this series, please go to: https://www.statforbiology.com/tags/drcte/. All these posts exapand on a paper that we have recently published in the Journal ‘Weed Science’; please follow this link to the paper.
A motivating ...
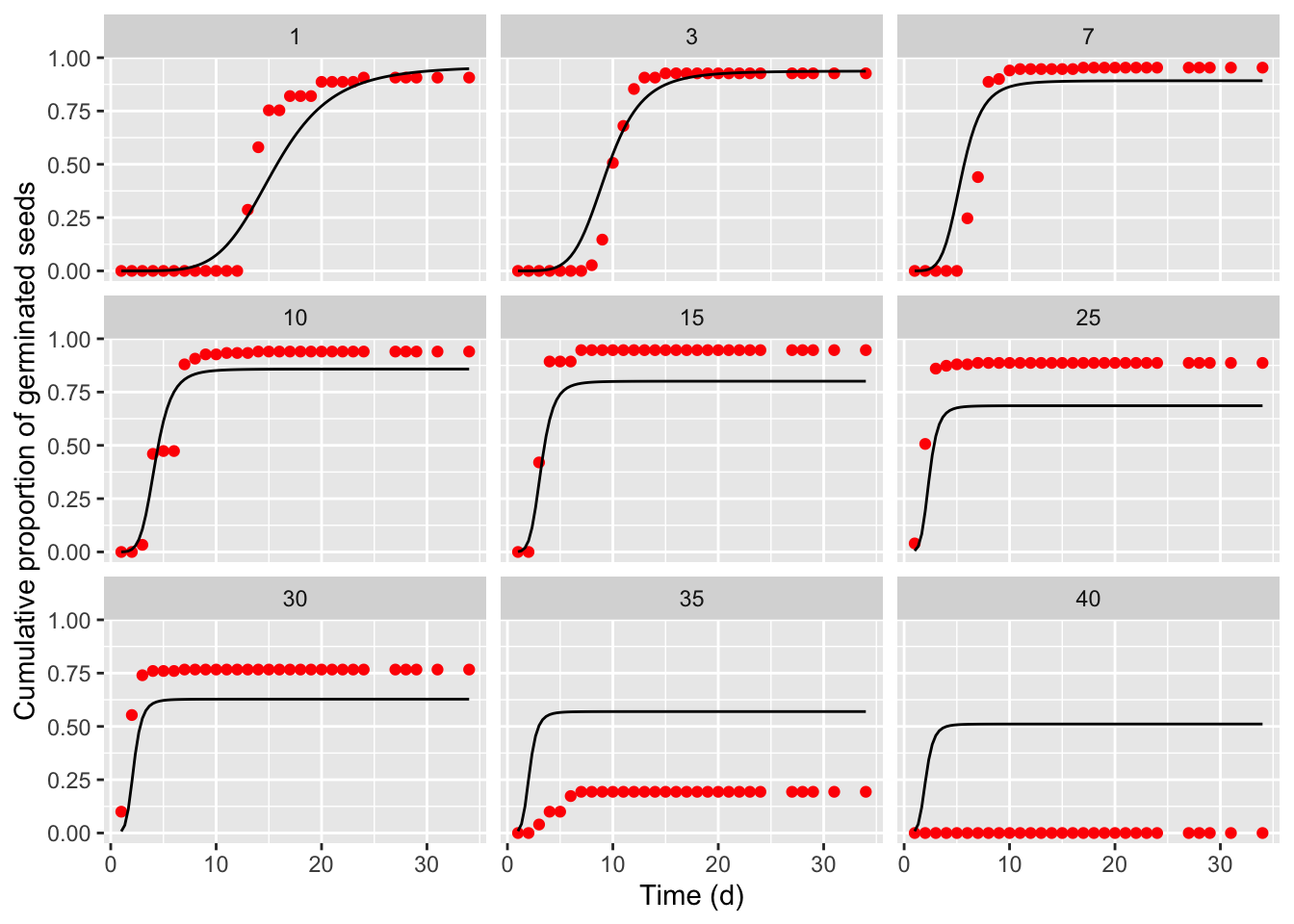
Fitting a hydro-thermal-time-model to seed germination data
This is a follow-up post. If you are interested in other posts of this series, please go to: https://www.statforbiology.com/tags/drcte/. All these posts exapand on a paper that we have recently published in the Journal ‘Weed Science’; please follow ...
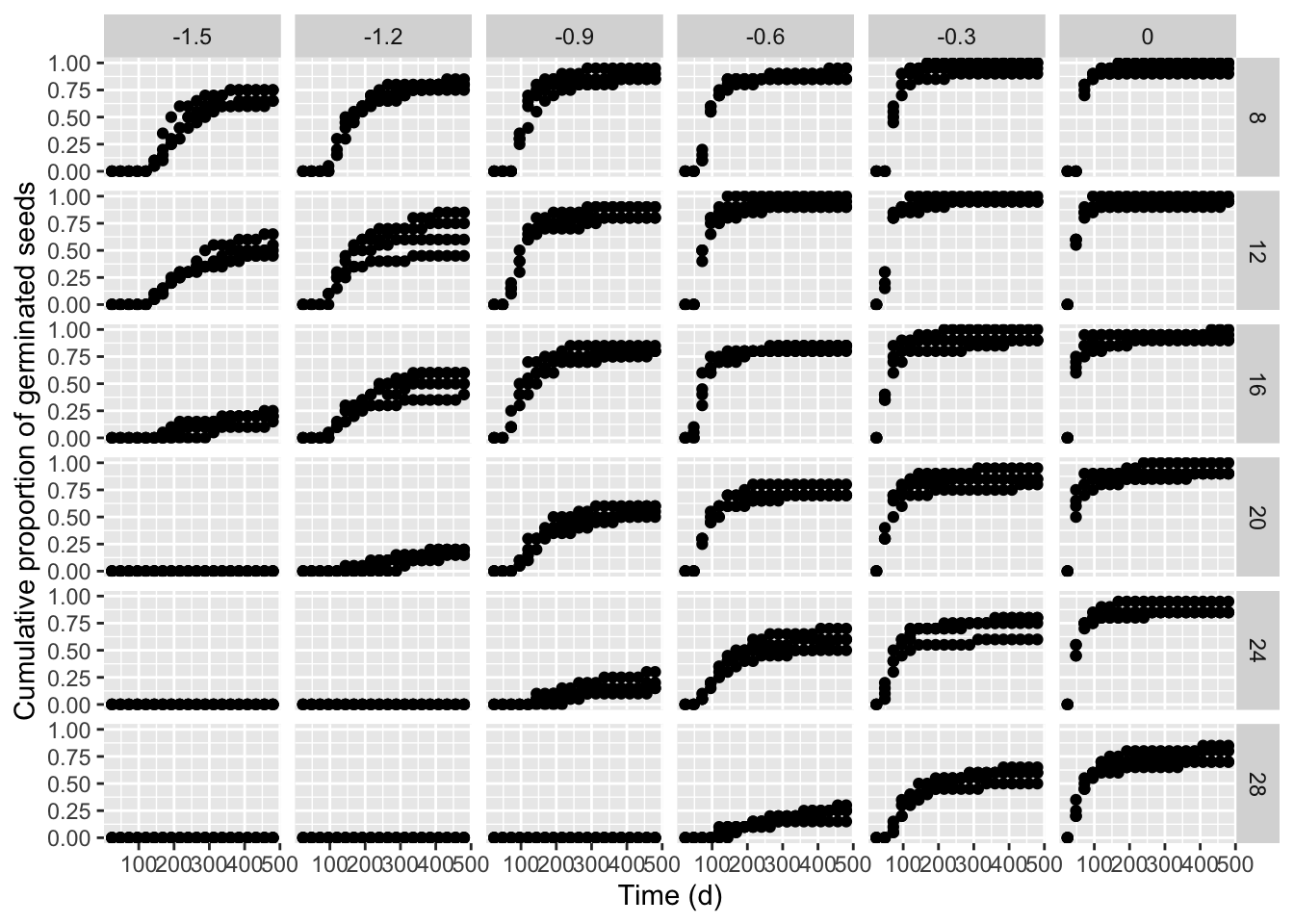
The coefficient of determination: is it the R-squared or r-squared?
We often use the coefficient of determination as a swift ‘measure’ of goodness of fit for our regression models. Unfortunately, there is no unique symbol for such a coefficient and both \(R^2\) and \(r^2\) are used in literature, almost interchangea... [Read more...]
Multi-environment split-plot experiments
Have you made a split-plot field experiment? Have you repeated such an experiment in two (or more) years/locations? Have you run into troubles, because the reviewer told you that your ANOVA model was invalid? If so, please, stop for awhile and read:... [Read more...]
Meta-analysis for a single study. Is it possible?
We all know that the word meta-analysis encompasses a body of statistical techniques to combine quantitative evidence from several independent studies. However, I have recently discovered that meta-analytic methods can also be used to analyse the re... [Read more...]
Should I say ”there is no difference” or ”the difference is not significant”?
In a recent manuscript we wrote a sentence similar to the following: “On average, the genotype A gave a yield of 12.4 tons per hectare, while the genotype B gave 10.6 tons per hectare and such a difference was not significant (P = 0.20)”. Perhaps I... [Read more...]
Analysing seed germination and emergence data with R (a tutorial). Part 9
This is a follow-up post. If you are interested in other posts of this series, please go to: https://www.statforbiology.com/tags/drcte/. All these posts expand on a manuscript that we have recently published in the Journal ‘Weed Science’; please follow this link to the paper. In order ...
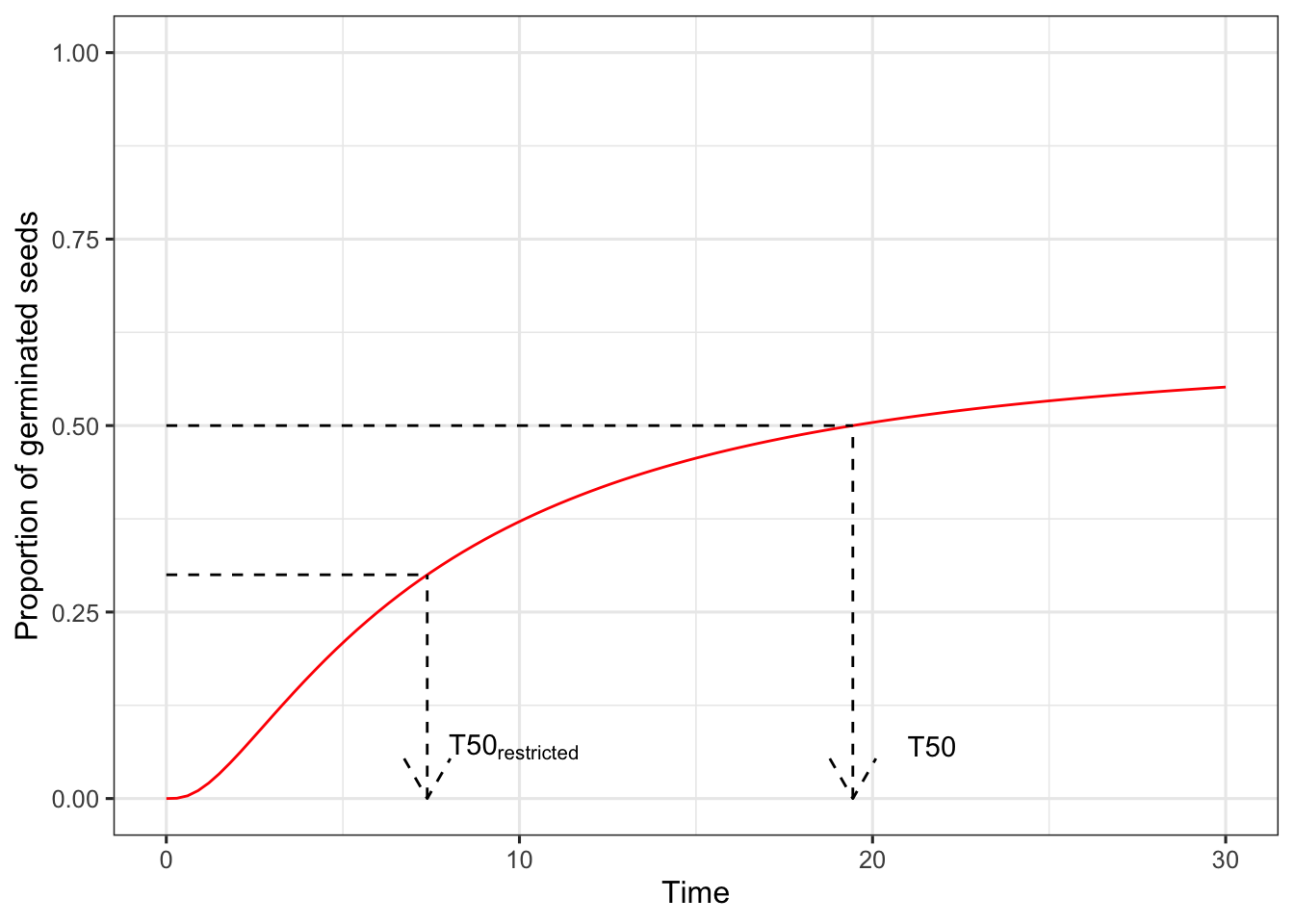
Analysing seed germination and emergence data with R (a tutorial). Part 8
This is a follow-up post. If you are interested in other posts of this series, please go to: https://www.statforbiology.com/tags/drcte/. All these posts expand on a paper that we have recently published in the Journal ‘Weed Science’; please follow this link to the paper.
Predictions from ... [Read more...]
Analysing seed germination and emergence data with R (a tutorial). Part 7
This is a follow-up post. If you are interested in other posts of this series, please go to: https://www.statforbiology.com/tags/drcte/. All these posts exapand on a paper that we have recently published in the Journal ‘Weed Science’; please follow this link to the paper.
Exploring the ... [Read more...]
Copyright © 2025 | MH Corporate basic by MH Themes