Understanding leaf node numbers when using rpart and rpart.rules
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
I recently ran into an issue with matching rules from a decision tree (output of rpart.plot::rpart.rules()) with leaf node numbers from the tree object itself (output of rpart::rpart()). This post explains the issue and how to solve it.
First, let’s build a decision tree model and print its tree representation:
library(rpart.plot) data(ptitanic) model <- rpart(survived ~ ., data = ptitanic, cp = .02) rpart.plot(model, extra = 101)
In the plot above, the two numbers in each node denote the number of observations in each class that fall into that node, while the percentage displayed is the percentage of all observations that fall into that node. This tree has 3 internal nodes and 4 leaves.
Row name in model$frame
The output of rpart() has a frame element which is a data frame with one row for each node in the tree (internal and external). The documentation (?rpart.object) says that “the row.names of frame contain the (unique) node numbers that follow a binary ordering indexed by node depth.” This will come in handy later. Here is part of the frame element for our tree:
cbind(model$frame[, 1:6], model$frame[,9][, 6]) # var n wt dev yval complexity model$frame[, 9][, 6] # 1 sex 1309 1309 500 1 0.424 1.00000000 # 2 age 843 843 161 1 0.021 0.64400306 # 4 <leaf> 796 796 136 1 0.000 0.60809778 # 5 sibsp 47 47 22 2 0.021 0.03590527 # 10 <leaf> 20 20 1 1 0.020 0.01527884 # 11 <leaf> 27 27 3 2 0.020 0.02062643 # 3 <leaf> 466 466 127 2 0.015 0.35599694
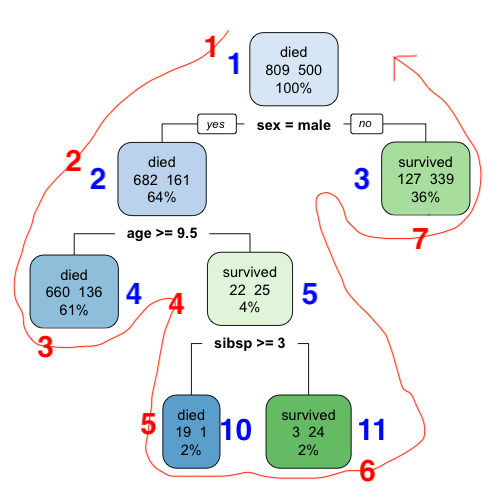
The slightly modified tree below shows how the row names match with the nodes. The orange numbers correspond to nodes that don’t actually exist in this tree: those would be the numbers for those nodes if the tree had nodes there.
The leaf nodes for this tree have row names 3, 4, 10 and 11.
Row number in model$frame
How does rpart() determine the order of the rows in model$frame? They are listed in preorder traversal order. Here is a visual description of that:
The numbers in red are the row numbers (notice how they go from 1 to 7 along the red line), while the numbers in blue are the row names. The leaf nodes for this tree have row numbers 3, 5, 6 and 7.
Leaf node number in model$where
The output from rpart() also has a where element that tells us which leaf node each observation in the dataset used to train the tree falls in. From the documentation, it “[contains] the row number of frame corresponding to the leaf node that each observation falls into.” In our context, the elements of frame would be one of {3, 5, 6, 7} (rather than one of {3, 4, 10, 11}).
head(model$where, n = 10) # 1 2 3 4 5 6 7 8 9 10 # 7 6 7 3 7 3 7 3 7 3
It’s easy to convert these leaf node row numbers into the leaf node row names:
head(row.names(model$frame)[model$where], n = 10) # [1] "3" "11" "3" "4" "3" "4" "3" "4" "3" "4"
Leaf node number in rpart.plot::rpart.rules()
The rpart.plot package has a function rpart.rules() that we can use to get the rules that define the leaf nodes as text strings:
rules <- rpart.rules(model) rules # survived # 0.05 when sex is male & age < 9.5 & sibsp >= 3 # 0.17 when sex is male & age >= 9.5 # 0.73 when sex is female # 0.89 when sex is male & age < 9.5 & sibsp < 3
The object returned by rpart.rules() might not be what you expect. It’s actually a data frame, where each column is part of the text string that you see printed above! The str() function makes this obvious:
str(rules) # Classes ‘rpart.rules’ and 'data.frame': 4 obs. of 13 variables: # $ survived: chr "0.05" "0.17" "0.73" "0.89" # $ : chr "when" "when" "when" "when" # $ : chr "sex" "sex" "sex" "sex" # $ : chr "is" "is" "is" "is" # $ : chr "male" "male" "female" "male" # $ : chr "&" "&" "" "&" # $ : chr "age" "age" "" "age" # $ : chr "< " ">=" "" "< " # $ : chr "9.5" "9.5" "" "9.5" # $ : chr "&" "" "" "&" # $ : chr "sibsp" "" "" "sibsp" # $ : chr ">=" "" "" "< " # $ : chr "3" "" "" "3" # - attr(*, "style")= chr "wide" # - attr(*, "eq")= chr "is" # - attr(*, "and")= chr "&" # - attr(*, "when")= chr "when"
Here is a view of the dataset in RStudio:
From this view, we can see that each row of the dataset has a name, and that name is the leaf node’s row name in frame (not the leaf node row number). If we want to, we can use these row names to match the rows here to the correct leaf nodes in frame.
Here is some code to transform the dataset object above into text strings, one for each node:
rule_strings <- apply(rules, 1, function(x) paste(x, collapse = " ")) rule_strings # 10 4 # "0.05 when sex is male & age < 9.5 & sibsp >= 3" "0.17 when sex is male & age >= 9.5 " # 3 11 # "0.73 when sex is female " "0.89 when sex is male & age < 9.5 & sibsp < 3"
Notice that this results in some extraneous white space for leaf nodes 3 and 4. The code below fixes that issue:
rule_strings <- apply(rules, 1, function(x) paste(x[x != ""], collapse = " ")) rule_strings # 10 4 # "0.05 when sex is male & age < 9.5 & sibsp >= 3" "0.17 when sex is male & age >= 9.5" # 3 11 # "0.73 when sex is female" "0.89 when sex is male & age < 9.5 & sibsp < 3"
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.