How much overdispersion is too much in typical GLMMs?
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
tl;dr: DHARMa tests will pick up on overdispersion before you see a rise of Type I error.
Overdispersion is a common problem in GL(M)Ms with fixed dispersion, such as Poisson or binomial GLMs. Here an explanation from the DHARMa vignette:
GL(M)Ms often display over/underdispersion, which means that residual variance is larger/smaller than expected under the fitted model. This phenomenon is most common for GLM families with constant (fixed) dispersion, in particular for Poisson and binomial models, but it can also occur in GLM families that adjust the variance (such as the beta or negative binomial) when distribution assumptions are violated.
The main issue with overdispersion is that
- p-values tend to be too small, thus leading to inflated Type I error
- CIs will be to small, thus leading to overconfidence about the precision of the estimate
Several R packages, notably DHARMa, allow testing GL(M)Ms for overdispersion. For version 0.3.4, we added a new parametric dispersion test, and we also recently ran a large number of additional comparative analysis on their power in different situation. The gist of this work is: the tests are pretty good at picking up on dispersion problems in a range of models.
But are those tests good enough? Or are they maybe too good, meaning that: if you get a significant dispersion test, should you switch to a variable dispersion glm (e.g. neg. binom), even if the dispersion problem is mild? To do a quick check of this question, I ran a few simulations using the DHARMa::runBenchmarks function.
library(DHARMa)
returnStatistics <- function(control = 1){
testData = createData(sampleSize = 200, family = poisson(),
overdispersion = control, fixedEffects = 0,
randomEffectVariance = 0)
fittedModel <- glm(observedResponse ~ Environment1, data = testData, family = poisson())
x = summary(fittedModel)
res <- simulateResiduals(fittedModel = fittedModel, n = 250)
out <- c("Type I error GLM slope" = x$coefficients[2,4],
"DHARMa testDispersion" = testDispersion(res, plot = FALSE)$p.value)
return(out)
}
out = runBenchmarks(returnStatistics, controlValues = seq(0, 1.5, 0.05), nRep = 500)
plot(out, xlab = "Added dispersion sd", ylab = "Prop significant", main = "n = 200")
The idea of these simulations is to slowly increase the overdispersion in a Poisson GLM where the true slope of a tested effect (Environment1) is zero. As overdispersion increases, we will get increasing Type I error (false positives) on the slope. We can then compare the Power of the DHARMa::testDispersion function with the rate of Type I errors, to see if DHARMa would have warned us early enough about the problem, or if the dispersion tests are maybe warning too early, i.e. before calibration issues in the p-values of the GLM get serious (see Fig. 1, calculated for different sampleSizes n)
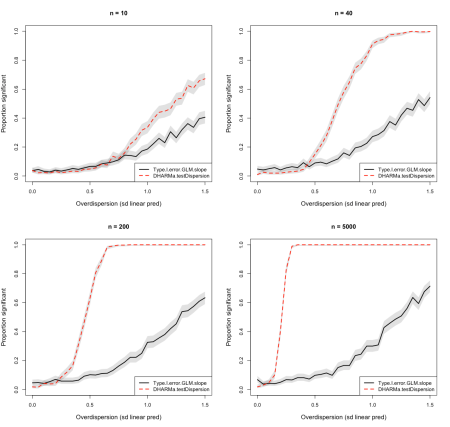
The results suggest that the power of DHARMa overdispersion tests depends more strongly on sample size than the increase of Type I error caused by the overdispersion. More precisely, for small sample sizes (n = 10/40), overdispersion tests pick up a signal roughly at the same point where overdispersion starts to create problems in the GLM estimates (i.e. false positives in regression effects).
For larger sample sizes (in particular n = 5000), however, even small levels of overdispersion are being picked up by DHARMa, whereas the GLM type I error is surprisingly unimpressed by the sample size. I have to say I was a bit surprised about the latter behaviour, and still do not fully understand it. It seems that the increase of type I error in a Poisson GLM mainly depends on the nominal dispersion and not so much on the sample size. Please comment if you have any idea about why this would be the case, I would have expected sample size to play a role as well.
Whatever the reason for the GLM behaviour, my conclusions (disclaimer: this is of course all only for a simple Poisson GLM, one should check if this generalises to other models) are as follows:
- In my simulations, problems with overdispersion were only substantial if a) tests are significant and b) the dispersion parameter is large, say e.g. > 2.
- This suggests that one could ignore significant overdispersion tests in DHARMa if n is large AND the estimated dispersion parameter is close to 1 (with the idea that the tests gest very sensitive for large n, thus picking up on minimal dispersion problems that are unproblematic for the GLMs)
- That being said: given that n is large, it seems no problem to support an additional parameter for adjusting the dispersion, e.g. through a negative binomial, so the question is if there is any reason to make this call
My overall recommendation would still be to move to a variable dispersion glm as soon as DHARMa dispersion tests flag overdispersion. But if you have particular reasons for avoiding this, you could ignore a positive test if n is large and the dispersion is close to 1.
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
