ppsr: An R implementation of the Predictive Power Score
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
A few months ago, I wrote about the Predictive Power Score (PPS): a handy metric to quickly explore and quantify the relationships in a dataset.
As a social scientist, I was taught to use a correlation matrix to describe the relationships in a dataset. Yet, in my opinion, the PPS provides three handy advantages:
- PPS works for any type of data, also nominal/categorical variables
- PPS quantifies non-linear relationships between variables
- PPS acknowledges the asymmetry of those relationships
Florian Wetschoreck came up with the PPS idea, wrote the original blog, and programmed a Python implementation of it (called ppscore).
Yet, I work mostly in R and I was very keen on incorporating this powertool into my general data science workflow.
So, over the holiday period, I did something I have never done before: I wrote an R package!
It’s called ppsr and you can find the code here on github.
Installation
# You can get the development version from GitHub:
# install.packages('devtools')
devtools::install_github('https://github.com/paulvanderlaken/ppsr')
Usage
The ppsr package has three main functions that compute PPS:
score()– which computes an x-y PPSscore_predictors()– which computes X-y PPSscore_matrix()– which computes X-Y PPS
Visualizing PPS
Subsequently, there are two main functions that wrap around these computational functions to help you visualize your PPS using ggplot2:
visualize_predictors()– producing a barplot of all X-y PPS

visualize_matrix()– producing a heatmap of all X-Y PPS

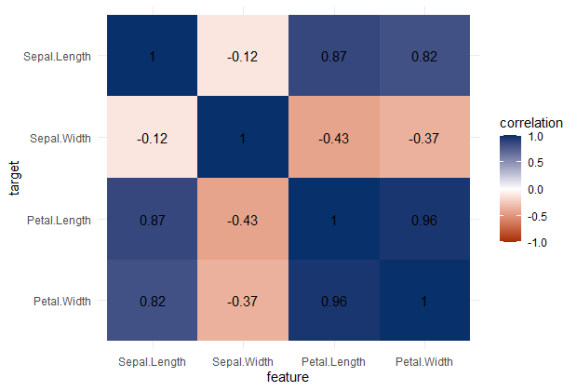
Note that Species is a nominal/categorical variable, with three character/text options.
A correlation matrix would not be able to show us that the type of iris Species can be predicted extremely well by the petal length and width, and somewhat by the sepal length and width. Yet, particularly sepal width is not easily predicted by the type of species.
Exploring mtcars
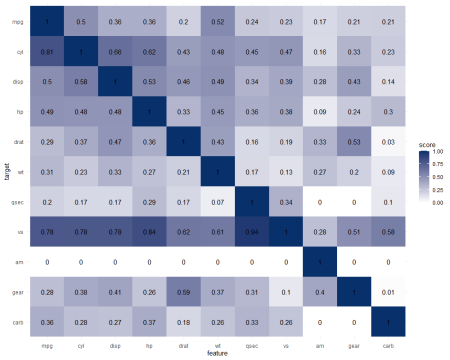
It takes about 10 seconds to run 121 decision trees with visualize_matrix(mtcars). Yet, the output is much more informative than the correlation matrix:
- cyl can be much better predicted by mpg than the other way around
- the classification of vs can be done well using nearly all variables as predictors, except for am
- yet, it’s hard to predict anything based on the vs classification
- a cars’ am can’t be predicted at all using these variables
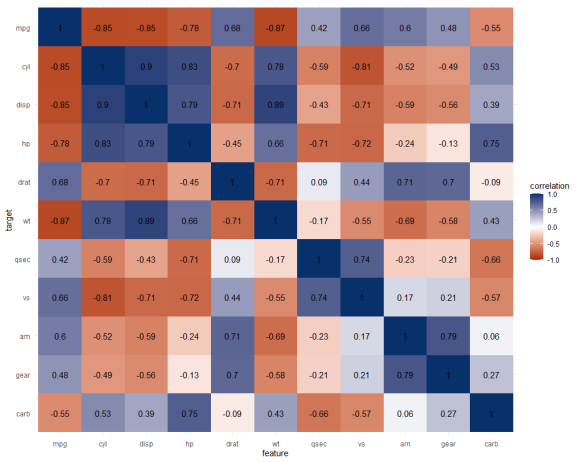
The correlation matrix does provides insights that are not provided by the PPS matrix. Most importantly, the sign and strength of any linear relationship that may exist. For instance, we can deduce that mpg relates strongly negatively with cyl.
Yet, even though half of the matrix does not provide any additional information (due to the symmetry), I still find it hard to derive the most important relations and insights at a first glance.
Moreover, the rows and columns for vs and am are not very informative in this correlation matrix as it contains pearson correlations coefficients by default, whereas vs and am are binary variables. The same can be said for cyl, gear and carb, which contain ordinal categories / integer data, so you can discuss the value of these coefficients depicted here.
Exploring trees
In R, there are many datasets built in via the datasets package. Let’s explore some using the ppsr::visualize_matrix() function.
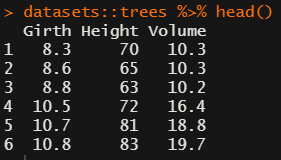
datasets::trees has data on 31 trees’ girth, height and volume.
visualize_matrix(datasets::trees) shows that both girth and volume can be used to predict the other quite well, but not perfectly.

Let’s have a look at the correlation matrix.
The scores here seem quite higher in general. A near perfect correlation between volume and girth.

Is it near perfect though? Let’s have a look at the underlying data and fit a linear model to it.
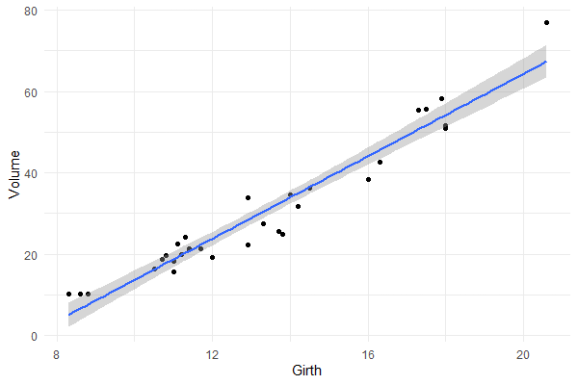
You will still be pretty far off the real values when you use a linear model based on Girth to predict Volume. This is what the original PPS of 0.65 tried to convey.
Actually, I’ve run the math for this linaer model and the RMSE is still 4.11. Using just the mean Volume as a prediction of Volume will result in 16.17 RMSE. If we map these RMSE values on a linear scale from 0 to 1, we would get the PPS of our linear model, which is about 0.75.
So, actually, the linear model is a better predictor than the decision tree that is used as a default in the ppsr package. That was used to generate the PPS matrix above.
Yet, the linear model definitely does not provide a perfect prediction, even though the correlation may be near perfect.
Conclusion
In sum, I feel using the general idea behind PPS can be very useful for data exploration.
Particularly in more data science / machine learning type of projects. The PPS can provide a quick survey of which targets can be predicted using which features, potentially with more complex than just linear patterns.
Yet, the old-school correlation matrix also still provides unique and valuable insights that the PPS matrix does not. So I do not consider the PPS so much an alternative, as much as a complement in the toolkit of the data scientist & researcher.
Enjoy the R package, or the Python module for that matter, and let me know if you see any improvements!
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
