Plot predicted values for presences vs. absences
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
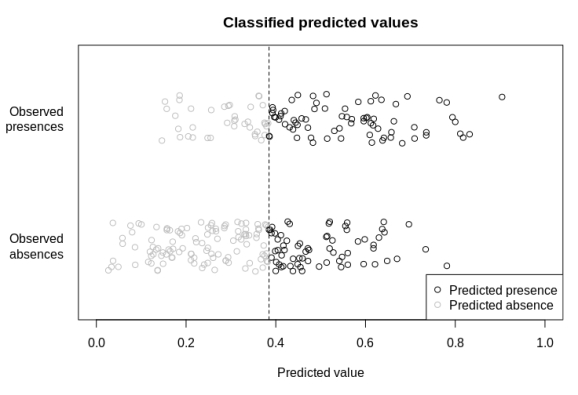
Diagnostic plots are always a nice, visually explicit way to assess our data and predictions, and I’ve just added a new one to the modEvA R package. The ‘predPlot‘ function plots predicted values separated into observed presences and absences, and coloured according to whether they’re above or below a given prediction threshold (the default threshold is species prevalence, i.e. proportion of presences, in the observed data). It shows whether there’s a visible split in the predicted values for presences and absences (see also functions predDensity and plotGLM in the same package, for alternative/complementary ways of visualizing this). The plot imitates (with permission from the author) one of the graphical outputs of the ‘summary‘ of models built with the ‘embarcadero‘ package (Carlson, 2020), but it can be applied to any ‘glm‘ object or any set of observed and predicted values, and it allows specifying a user-defined prediction threshold. The ‘predPlot‘ function is now included in package modEvA (version >= 2.1, currently available on R-Forge).
predPlot <- function(model = NULL, obs = NULL, pred = NULL, thresh = "preval", main = "Classified predicted values", legend.pos = "bottomright") {
# version 1.0 (20 Jan 2021)
if (!is.null(model)) {
if (!("glm" %in% class(model)) || family(model)$family != "binomial") stop("'model' must be of class 'glm' and family 'binomial'.")
if (!is.null(obs)) message("Argument 'obs' ignored in favour of 'model'.")
if (!is.null(pred)) message("Argument 'pred' ignored in favour of 'model'.")
obs <- model$y
pred <- model$fitted.values
} else {
if (is.null(obs) || is.null(pred)) stop ("You must provide either 'model' or a combination of 'obs' and 'pred'.")
if (!is.numeric(obs) || !is.numeric(pred)) stop ("'obs' and 'pred' must be numeric.")
if (length(obs) != length(pred)) stop("'obs' and 'pred' must have the same length.")
}
if (!(thresh == "preval" || (is.numeric(thresh) && thresh >= 0 && thresh <= 1))) stop ("'thresh' must be either 'preval' or a numeric value between 0 and 1.")
if (thresh == "preval") thresh <- prevalence(obs)
pred0 <- pred[obs == 0]
pred1 <- pred[obs == 1]
opar <- par(no.readonly = TRUE)
on.exit(par(opar))
par(mar = c(5, 5.2, 3, 1))
plot(x = c(0, 1), y = c(-0.5, 1.5), xlab = "Predicted value", type = "n", ylab = "", yaxt = "n", main = main)
axis(side = 2, at = c(0, 1), tick = FALSE, labels = c("Observed\nabsences", "Observed\npresences"), las = 1)
abline(v = thresh, lty = 2)
points(x = pred0, y = sapply(rep(0, length(pred0)), jitter, 10), col = ifelse(pred0 < thresh, "grey", "black"))
points(x = pred1, y = sapply(rep(1, length(pred1)), jitter, 10), col = ifelse(pred1 < thresh, "grey", "black"))
if (!is.na(legend.pos) && legend.pos != "n") legend(legend.pos, legend = c("Predicted presence", "Predicted absence"), pch = 1, col = c("black", "grey"))
}
Usage examples:
library(modEvA) # load sample models: data(rotif.mods) # choose a particular model to play with: mod <- rotif.mods$models[[1]] # make some predPlots: predPlot(model = mod) predPlot(model = mod, thresh = 0.5) # you can also use 'predPlot' with vectors of observed and predicted values instead of a model object: myobs <- mod$y mypred <- mod$fitted.values predPlot(obs = myobs, pred = mypred)
References
Carlson C.J. (2020) embarcadero: Species distribution modelling with Bayesian additive regression trees in R. Methods in Ecology and Evolution, 11: 850-858.
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
