Downloading food web databases and deriving basic structural metrics
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
This blog post presents a beta version of the package to download, process and derive metrics from food web datasets.
DISCLAIMER: This is an early release. The code will be subject to improvements with time. Some of this code has been adapted from other sources (e.g. Ecobase website). A few errors are expected when running these functions. Any suggestions are welcomed! Plenty of room for improvement…
Additional information on the package is available on GitHub.
First, we need to install Load required packages:
install_github("FMestre1/fw_package")
Create food web list (from the mangal database as an example):
#mg1 <- create.fw.list(db="mg", ref=TRUE, spatial = TRUE)
#(this takes a long time...)
#To make it faster, let's just upload the data:
data(mg1)
Which of the matrices are adjacency matrices (0 and 1 matrices)?
is.adjacency.matrix(mg1) ## Matrix 1 is an adjacency matrix! ## Matrix 2 is an adjacency matrix! ## Matrix 3 is an adjacency matrix! ## Matrix 4 is an adjacency matrix! ## Matrix 5 is an adjacency matrix! (...) ## Matrix 387 is an adjacency matrix! ## Matrix 388 is an adjacency matrix! ## Matrix 389 is an adjacency matrix! ## Matrix 390 is an adjacency matrix!
Need to convert to adjacency matrix (not needed here)?
mg2 <- convert2adjacency(mg1) is.adjacency.matrix(mg2) ## Matrix 1 is an adjacency matrix! ## Matrix 2 is an adjacency matrix! ## Matrix 3 is an adjacency matrix! ## Matrix 4 is an adjacency matrix! ## Matrix 5 is an adjacency matrix! (...) ## Matrix 386 is an adjacency matrix! ## Matrix 387 is an adjacency matrix! ## Matrix 388 is an adjacency matrix! ## Matrix 389 is an adjacency matrix! ## Matrix 390 is an adjacency matrix!
Which are square matrices (having the same number of columns and rows)?
is.sq.matrix(mg1) ## Matrix 1 already is square! ## Matrix 2 already is square! ## Matrix 3 already is square! ## Matrix 4 already is square! ## Matrix 5 already is square! (...) ## Matrix 385 already is square! ## Matrix 386 already is square! ## Matrix 387 already is square! ## Matrix 388 already is square! ## Matrix 389 already is square! ## Matrix 390 already is square!
Need to convert to square matrix (not needed here)?
mg3 <- rect2square(mg2) ## Matrix 1 already is square! ## Matrix 2 already is square! ## Matrix 3 already is square! ## Matrix 4 already is square! ## Matrix 5 already is square! (...) ## Matrix 388 already is square! ## Matrix 389 already is square! ## Matrix 390 already is square!
Having the full dataset, with all the matrices with the same format, now we will derive network metrics for each food web matrix (number of nodes, number of trophic interactions, linkage density, connectance, compartmentalization and maximum trophic level).
metrics <- fw.metrics(mg1) ## Computing network metrics for food web 1 of 390... ## Computing network metrics for food web 2 of 390... ## Computing network metrics for food web 3 of 390... ## Computing network metrics for food web 4 of 390... ## Computing network metrics for food web 5 of 390... names(metrics) ## [1] "number_nodes" "number_links" "link_density" ## [4] "connectance" "compartmentalization" "maximum_trophic_level"
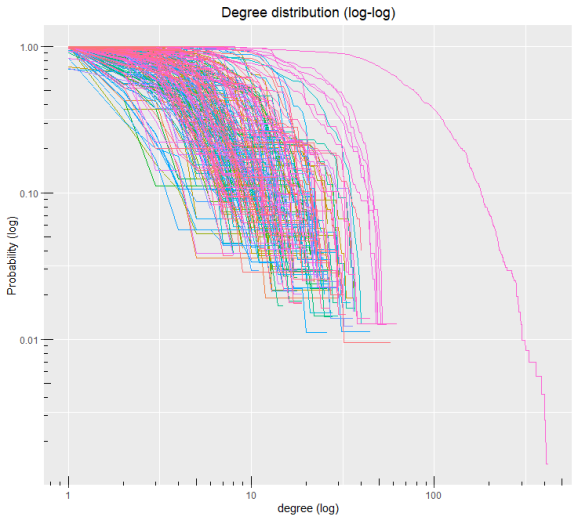
Finally, we can plot the degree distribution of all food webs in the dataset:
dd.fw(mg1, log=TRUE, cumulative=TRUE)
## iter degree1 probability ## 1 iter1 1 0.98484848 ## 2 iter1 2 0.83333333 ## 3 iter1 3 0.75757576 ## 4 iter1 4 0.62121212 ## 5 iter1 5 0.53030303
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
