Veterinary Epidemiologic Research: Linear Regression Part 3 – Box-Cox and Matrix Representation
[This article was first published on denis haine » R, and kindly contributed to R-bloggers]. (You can report issue about the content on this page here)
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
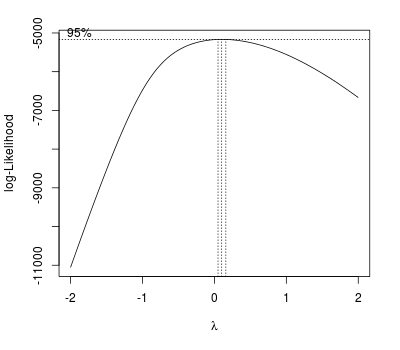
In the previous post, I forgot to show an example of Box-Cox transformation when there’s a lack of normality. The Box-Cox procedure computes values of which best “normalises” the errors.
| |
Transformed value of Y |
|---|---|
| 2 | |
| 1 | |
| 0.5 | |
| 0 | |
| -0.5 | |
| -1 | |
| -2 | |
For example:
lm.wpc2 <- lm(wpc ~ vag_disch + milk120 + milk120.sq, data = daisy3) library(MASS) boxcox(lm.wpc2, plotit = TRUE)
The plot indicates a log transformation.
Matrix Representation
We can use a matrix representation of the regression equation. The regression equation can be written as
where
,
,
and
.
With the following regression:
lm.wpc3 <- lm(wpc ~ milk120 + hs100_ct, data = daisy3)
(lm.wpc3.sum <- summary(lm.wpc3))
Call:
lm(formula = wpc ~ milk120 + hs100_ct, data = daisy3)
Residuals:
Interval from wait period to conception
Min 1Q Median 3Q Max
-71.83 -36.57 -15.90 23.86 211.18
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 75.884063 6.115894 12.408 <2e-16 ***
milk120 -0.001948 0.001858 -1.049 0.294
hs100_ct 18.530340 2.106920 8.795 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 50.53 on 1522 degrees of freedom
Multiple R-squared: 0.0495, Adjusted R-squared: 0.04825
F-statistic: 39.63 on 2 and 1522 DF, p-value: < 2.2e-16
anova(lm.wpc3)
Analysis of Variance Table
Response: wpc
Df Sum Sq Mean Sq F value Pr(>F)
milk120 1 4865 4865 1.9053 0.1677
hs100_ct 1 197512 197512 77.3518 <2e-16 ***
Residuals 1522 3886309 2553
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
We start by making the X matrix and response :
X <- cbind(1 , daisy3[, c(8, 24)]) # or alternatively: model.matrix(lm.wpc3) y <- daisy3$wpc
Then we get the :
XX <- t(X) %*% X
# or alternatively: crossprod(X, X)
XX
1 milk120 hs100_ct
1 1525.00000 4907834.4 -29.87473
milk120 4907834.39954 16535517546.0 -120696.45468
hs100_ct -29.87473 -120696.5 576.60900
We calculate the inverse of :
XX.inv <- solve(t(X) %*% X)
# or alternatively: qr.solve(XX)
# or also: lm.wpc3.sum$cov.unscaled
XX.inv
1 milk120 hs100_ct
1 1.464864e-02 -4.348902e-06 -1.513557e-04
milk120 -4.348902e-06 1.351675e-09 5.761289e-08
hs100_ct -1.513557e-04 5.761289e-08 1.738495e-03
Now to get the coefficients estimates:
XX.inv %*% t(X) %*% y
# or alternatively: solve(t(X) %*% X, t(X) %*% y)
# or also: XX.inv %*% crossprod(X, y)
# or: qr.solve(X, y)
[,1]
1 75.884062540
milk120 -0.001948437
hs100_ct 18.530340385
The fitted values:
crossprod(t(X), qr.solve(X, y)
SSE and MSE;
SSE <- crossprod(y, y) - crossprod(y, yhat)
SSE
[,1]
[1,] 3886309
MSE <- SSE / (length(y) - 3)
MSE
[,1]
[1,] 2553.423
The residual standard error:
sqrt(sum(lm.wpc3$res^2) / (1525 - 3))
The standard errors of the coefficients and the :
sqrt(diag(XX.inv)) * 50.5314
1 milk120 hs100_ct
6.115893490 0.001857794 2.106920139
1 - sum(lm.wpc3$res^2) / sum((y - mean(y))^2)
[1] 0.04949679
To leave a comment for the author, please follow the link and comment on their blog: denis haine » R.
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.