discrimination between CpG islands and random sequences using Markov chains
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
Major part of modern research is trying to find patterns in the given dataset using learning methods. One of the methods that can use a priori information for such purpose is Markov chains, in which the probability of symbol occurrence depends only on previous symbol (appropriate for the dinucleotide modeling).
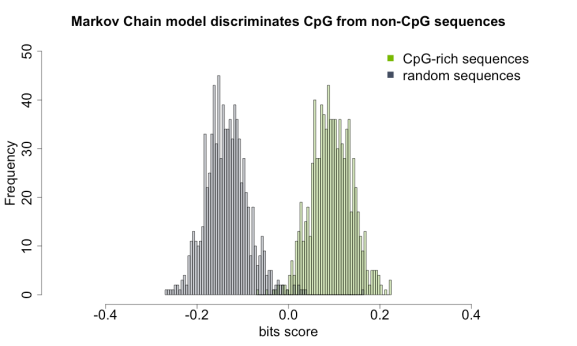
The question I am addressing here is – given the random sequence from mouse genome, can you predict that this is a CpG island? In the next few lines of code you will find many useful approaches to MC modeling, splitting of odd and even elements of R vector using “substring” function, querying the UCSC database solely via R, and informative visualization with transparent histograms.
Note that I got sequences from BSgenome.Mmusculus.UCSC.mm9 library, which contains complete mouse genome. Since the download of the same might be tedious for some (~750mb), I uploaded the cpg and random regions to pastebin.
CpG Islands – http://pastebin.com/372cfs6U
Random – http://pastebin.com/wZyKFFkx
# load libraries
library(compiler)
library(rtracklayer)
library(GenomicFeatures)
# ignore following if you're taking sequences from pastebin
library(BSgenome.Mmusculus.UCSC.mm9)
# models derived by ML estimator (a_st = c_st / sum(c_st')) from CpG and non-CpG islands
# in each row, nucleotide has a probability of being followed by respective base in the column,
# thus rows should sum up to 1
# notice the drop of G being followed by C in negative model (due to C being target of spontaneous deamination upon methylation),
# same is preserved in CpG-islands rich regions due to the lack of methylation supression on promoter regions
positive.model <- t(matrix(
data = c(0.180, 0.274, 0.426, 0.120, 0.171, 0.368, 0.274, 0.188, 0.161, 0.339, 0.375, 0.125, 0.079, 0.355, 0.384, 0.182),
nrow = 4,
ncol = 4,
dimnames = list(c("A", "C", "G", "T"), c("A", "C", "G", "T"))
))
negative.model <- t(matrix(
data = c(0.300, 0.205, 0.285, 0.210, 0.322, 0.298, 0.078, 0.302, 0.248, 0.246, 0.298, 0.208, 0.177, 0.239, 0.292, 0.292),
nrow = 4,
ncol = 4,
dimnames = list(c("A", "C", "G", "T"), c("A", "C", "G", "T"))
))
# log-likelihood table derived from log-odds ratio of two models
# used for discrimination of sequences as CpG or non-CpG islands
likelihood.values <- round(log2(positive.model/negative.model), 3)
# score (in bits) of the sequence normalized by its length
bits.score <- function(sequence = DNA_region, model = likelihood.values) {
# sequence length
seq.length <- nchar(sequence)
# if sequence is odd, trim the last element
if (seq.length %% 2 != 0) {
sequence <- paste(strsplit(sequence, split = "")[[1]][-seq.length], collapse = "")
seq.length <- seq.length - 1
}
# extract dinucleotides
idx <- seq(1, seq.length)
odds <- idx[(idx %% 2) == 1]
evens <- idx[(idx %% 2) == 0]
tmp.split <- substring(sequence, odds, evens)
# return normalized sum of the log-likelihood scores
return(sum(unlist(lapply(tmp.split, function(tmp.dinucleotide) {
lookup <- strsplit(tmp.dinucleotide, split = "")[[1]]
return(model[lookup[1], lookup[2]])
}))) / seq.length)
}
bits.score <- cmpfun(bits.score)
# setting up the discrimination model
model.score <- function(sequences = DNA_region) {
all.dinucleotides <- do.call("c", lapply(sequences, function(sequence) {
# sequence length
seq.length <- nchar(sequence)
# if sequence is odd, trim the last element
if (seq.length %% 2 != 0) {
sequence <- paste(strsplit(sequence, split = "")[[1]][-seq.length], collapse = "")
seq.length <- seq.length - 1
}
# extract dinucleotides
idx <- seq(1, seq.length)
odds <- idx[(idx %% 2) == 1]
evens <- idx[(idx %% 2) == 0]
tmp.split <- substring(sequence, odds, evens)
}))
all.counts <- t(matrix(table(all.dinucleotides), ncol = 4, nrow = 4, dimnames = list(c("A", "C", "G", "T"), c("A", "C", "G", "T"))))
all.counts <- t(apply(all.counts, 1, function(tmp.row) { tmp.row / sum(tmp.row) } ))
return(all.counts)
}
model.score <- cmpfun(model.score)
# following code extracts sequences from annotated and random parts of the mm9 genome
# ignore it if you're using pastebin text
# querying the UCSC database for CpG islands track
session <- browserSession()
genome(session) <- "mm9"
query <- ucscTableQuery(session, "CpG Islands")
cpg.islands <- getTable(query)
cpg.islands <- subset(cpg.islands, cpg.islands$chrom == "chr1", select = c("chrom", "chromStart", "chromEnd", "length"))
colnames(cpg.islands) <- c("chr", "region.start", "region.end", "region.width")
# generate random positions via "sample" function, but not exceeding largest width of CpG island
random.regions <- data.frame(chr = "chr1", region.start = sample(seq(0, seqlengths(Mmusculus)[["chr1"]] - max(cpg.islands$region.width)), nrow(cpg.islands)))
random.regions$region.end <- random.regions$region.start + cpg.islands$region.width
markov.chain.seq <- lapply(c("cpg.islands", "random.regions"), FUN = function(tmp.sample) {
tmp.dataset <- get(tmp.sample)
if (nrow(tmp.dataset) > 0) {
tmp.sview <- as.character(DNAStringSet(Views(unmasked(Mmusculus[["chr1"]]), start = tmp.dataset$region.start, end = tmp.dataset$region.end)))
}
return(tmp.sview)
})
# only keep sequences from non-repetitive parts of genome
markov.chain.seq <- lapply(markov.chain.seq, function(tmp.sample) { do.call("c", lapply(tmp.sample, function(tmp.seq) { tmp.seq[!grepl("N", tmp.seq)] })) })
names(markov.chain.seq) <- c("cpg.islands", "random.regions")
# you can substitute "lapply" with "mclapply" for parallel processing with multicore package at this point
# output of the discrimination plot
png("~/MarkovChain.png", 1000, 600)
hist(unlist(lapply(markov.chain.seq[[1]], bits.score)), breaks = 100, xlim = c(-0.5, 0.5), ylim = c(0, 50), col = "#78B90050", main = "Markov Chain model discriminates CpG from non-CpG sequences", xlab = "bits score")
hist(unlist(lapply(markov.chain.seq[[2]], bits.score)), breaks = 100, col = "#444D6150", add = TRUE)
legend("topright", c("CpG-rich sequences", "random sequences"), col = c("#78B900", "#444D61"), pch = 15, cex = 2, bty = "n")
dev.off()
Similar to the HMM example from previous post, the Markov chain is also based on Durbin (1998.) book.
Cheers, mintgene.
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.