Monster functions (Raaargh!)
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
It’s widely considered good programming practice to have lots of little functions rather than a few big functions. The reasons behind this are simple. When your program breaks, it’s much nicer to debug a five line function than a five hundred line function. Additionally, by breaking up your code into little chunks, you often find that some of those chunks are reusable in other contexts, saving you re-writing code in your next project. The process of breaking your code down into these smaller chunks is called refactoring.
The concept of a line of code is surprisingly fluid in R. Since you can add whitespace more or less where you like, the same code can take up one line in your editor of hundreds, if you so choose. Assuming that most programmers will write in a reasonably standard way, we can get a rough idea of how many lines there are in an R function by calling deparse on its body. deparse is less scary than it sounds. Parsing means turning a load of text into something meaningful; thus deparsing means turning something meaningful into a load of text. deparse essentially works like as.character for expressions. (Actually, you can call as.character on expressions, but the results are often dubious.)
A very interesting question is “how much of base R could do with refactoring into smaller pieces?”. To answer this, our first task is to get all the functions.
fn_names <- apropos(".+")
fns <- lapply(fn_names, get)
names(fns) <- fn_names
fns <- Filter(is.function, fns)
apropos finds all the functions on your search path (i.e., from all the packages that have been loaded). Try this code with a freshly loaded version of R, and again with all your packages loaded. The function below will do that for you.
load_all_packages <- function()
{
invisible(sapply(
rownames(installed.packages()),
require,
character.only = TRUE
))
}
load_all_packages()
The number of lines in each function is very straightforward to get from here.
n_lines_in_body <- function(fn)
{
length(deparse(body(fn)))
}
n_lines <- sapply(fns, n_lines_in_body)
Let’s take a look at the distribution of those lengths.
library(ggplot2) hist_line_count <- ggplot(data.frame(n_lines = n_lines), aes(n_lines)) + geom_histogram(binwidth = 5) hist_line_count
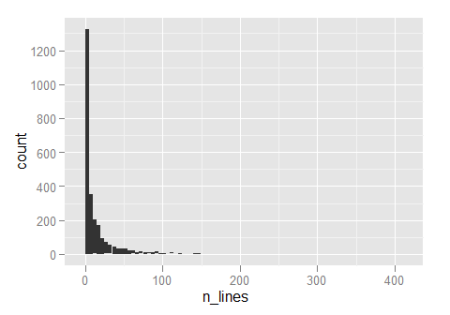
So about half the functions are five lines or less, which is all well and good. Notice that the x-axis extends all the way over to 400 though, so there clearly are some monsters in there.
head(sort(n_lines, decreasing = TRUE))
library arima help.search coplot loadNamespace plot.lm
409 328 320 316 305 299
So library is the number one culprit for being over long and complicated. In fairness to it though, it does mostly consist of sub-functions, so there clearly has been a lot of refactoring done on it; its just that the individual bits are contained within it rather than elsewhere. arima is more of a mess; it looks like the code is so old that no-one dare touch it anymore. None of these functions are really bad though. To see a package that really need some refactoring work, load up Hmisc and rerun this analysis. Now eight of the top ten longest functions come from this package. Quick challenge for you: hunting through other packages, can you find a function that beats Hmisc’s transcan at 591 lines?
Tagged: r, refactoring
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
