General Regression Neural Network with R
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
Similar to the back propagation neural network, the general regression neural network (GRNN) is also a good tool for the function approximation in the modeling toolbox. Proposed by Specht in 1991, GRNN has advantages of instant training and easy tuning. A GRNN would be formed instantly with just a 1-pass training with the development data. In the network development phase, the only hurdle is to tune the hyper-parameter, which is known as sigma, governing the smoothness of a GRNN.
The grnn package (http://flow.chasset.net/r-grnn/) is the implementation of GRNN in R and was just published on CRAN last month. Although the grnn package is still in the early phase, e.g. version 0.1, it is very easy to use and has a great potential for future improvements. For instance, the guess() function to predict new cases is only able to take 1 record at a time. Therefore, the user needs to write his / her own function to generate predicted values from a data frame. In addition, there is no automatic scheme to find the optimal value of the smooth parameter sigma. The user has to come up with his / her own solution.
Below is my test drive of grnn package over the weekend. By leveraging the power of foreach package, I wrote a simple function to let the guess() function able to score a whole matrix instead of a single row. Additionally, I used a hold-out sample to search for the optimal value of sigma, which turns out to work out pretty well and identifies the lowest SSE for the hold-out sample with sigma = 0.55.
pkgs <- c('MASS', 'doParallel', 'foreach', 'grnn')
lapply(pkgs, require, character.only = T)
registerDoParallel(cores = 8)
data(Boston)
# PRE-PROCESSING DATA
X <- Boston[-14]
st.X <- scale(X)
Y <- Boston[14]
boston <- data.frame(st.X, Y)
# SPLIT DATA SAMPLES
set.seed(2013)
rows <- sample(1:nrow(boston), nrow(boston) - 200)
set1 <- boston[rows, ]
set2 <- boston[-rows, ]
# DEFINE A FUNCTION TO SCORE GRNN
pred_grnn <- function(x, nn){
xlst <- split(x, 1:nrow(x))
pred <- foreach(i = xlst, .combine = rbind) %dopar% {
data.frame(pred = guess(nn, as.matrix(i)), i, row.names = NULL)
}
}
# SEARCH FOR THE OPTIMAL VALUE OF SIGMA BY THE VALIDATION SAMPLE
cv <- foreach(s = seq(0.2, 1, 0.05), .combine = rbind) %dopar% {
grnn <- smooth(learn(set1, variable.column = ncol(set1)), sigma = s)
pred <- pred_grnn(set2[, -ncol(set2)], grnn)
test.sse <- sum((set2[, ncol(set2)] - pred$pred)^2)
data.frame(s, sse = test.sse)
}
cat("\n### SSE FROM VALIDATIONS ###\n")
print(cv)
jpeg('grnn_cv.jpeg', width = 800, height = 400, quality = 100)
with(cv, plot(s, sse, type = 'b'))
cat("\n### BEST SIGMA WITH THE LOWEST SSE ###\n")
print(best.s <- cv[cv$sse == min(cv$sse), 1])
# SCORE THE WHOLE DATASET WITH GRNN
final_grnn <- smooth(learn(set1, variable.column = ncol(set1)), sigma = best.s)
pred_all <- pred_grnn(boston[, -ncol(set2)], final_grnn)
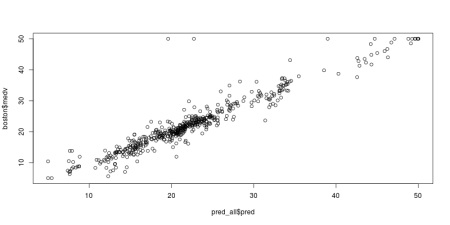
jpeg('grnn_fit.jpeg', width = 800, height = 400, quality = 100)
plot(pred_all$pred, boston$medv)
dev.off()
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.