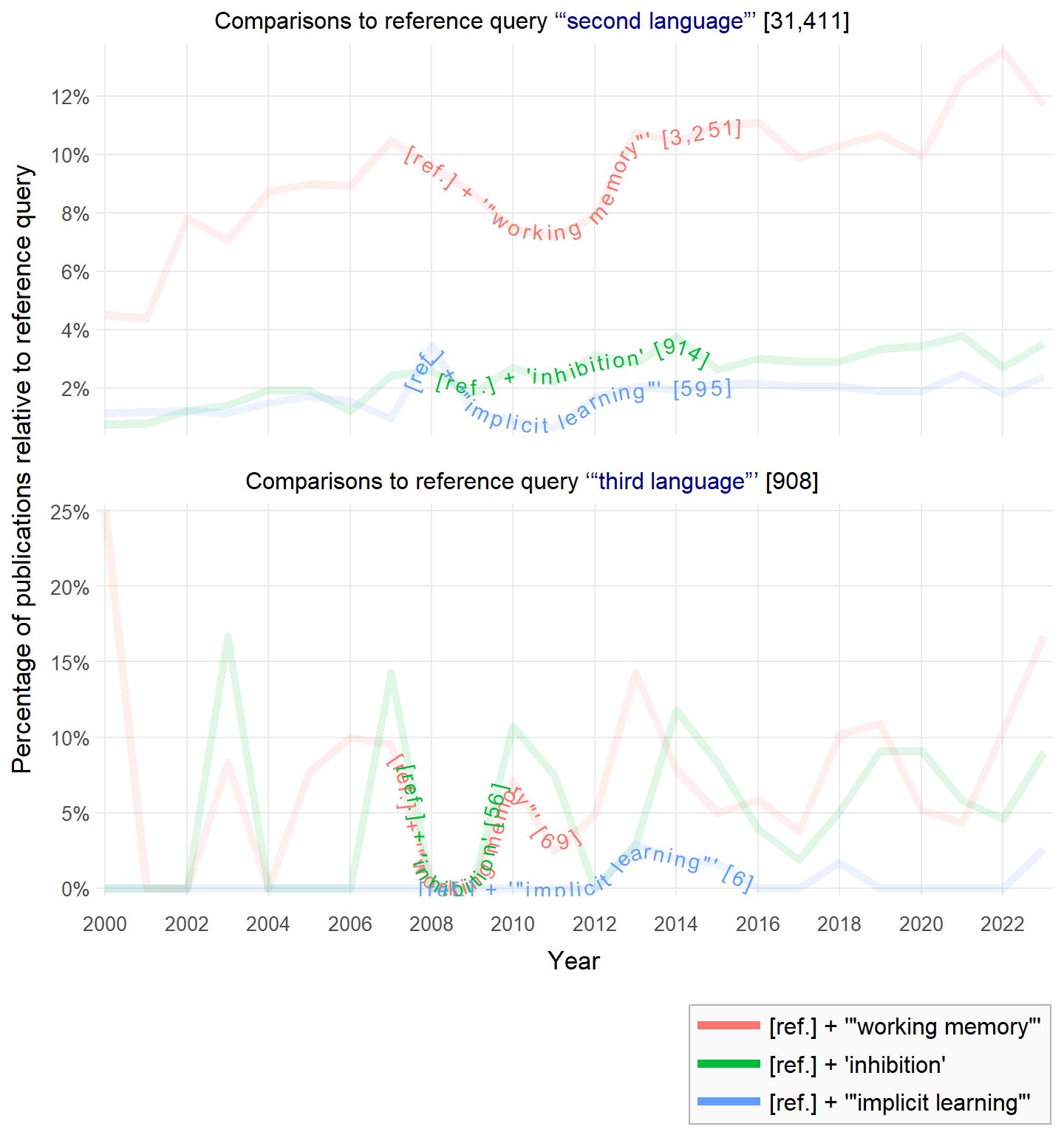
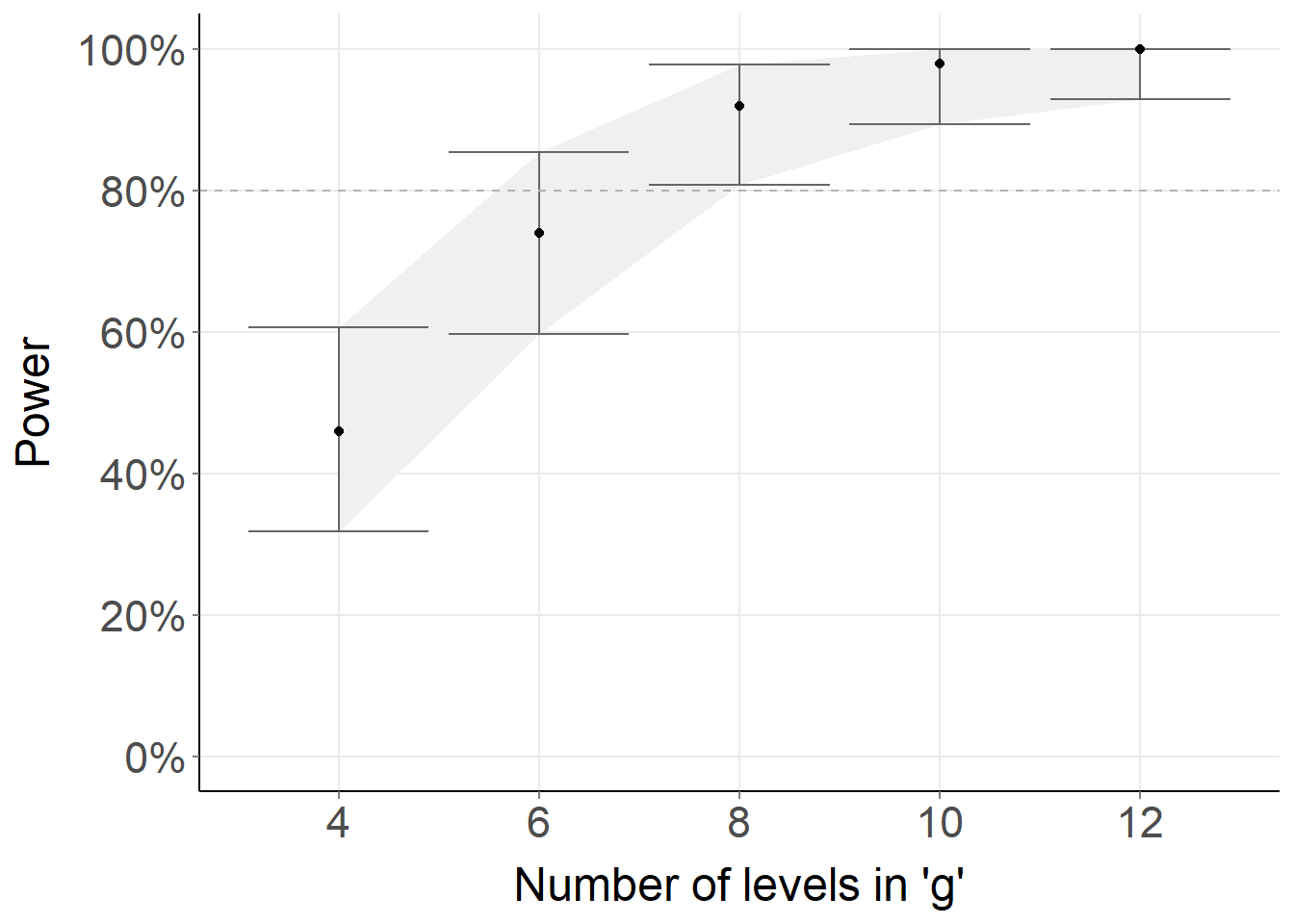
rscopus_plus: An extension of the rscopus package


library(lme4)
#> Loading required package: Matrix
library(sjPlot)
library(ggplot2)
theme_set(theme_sjplot())
# Create data using code by Ben Bolker from
# https://stackoverflow.com/a/38296264/7050882
set.seed(101)
spin = runif(600, 1, 24)
reg = runif(600, 1, 15)
ID = rep(c("1","2","3","4","5", "6", "7", "8", "9", "10"))
day = rep(1:30, each = 10)
testdata <- data.frame(spin, reg, ID, day)
testdata$fatigue <- testdata$spin * testdata$reg/10 * rnorm(30, mean=3, sd=2)
fit = lmer(fatigue ~ spin * reg + (1|ID),
data = testdata, REML = TRUE)
plot_model(fit, type = 'pred', terms = c('spin', 'reg'))
#> Warning: Ignoring unknown parameters: linewidth
...
library(lme4)
#> Loading required package: Matrix
library(sjPlot)
#> Learn more about sjPlot with 'browseVignettes("sjPlot")'.
library(ggplot2)
theme_set(theme_sjplot())
# Create data partially based on code by Ben Bolker
# from https://stackoverflow.com/a/38296264/7050882
set.seed(101)
spin = runif(800, 1, 24)
trait = rep(1:40, each = 20)
ID = rep(1:80, each = 10)
testdata <- data.frame(spin, trait, ID)
testdata$fatigue <-
testdata$spin * testdata$trait /
rnorm(800, mean = 6, sd = 2)
# Model
fit = lmer(fatigue ~ spin * trait + (1|ID),
data = testdata, REML = TRUE)
#> boundary (singular) fit: see help('isSingular')
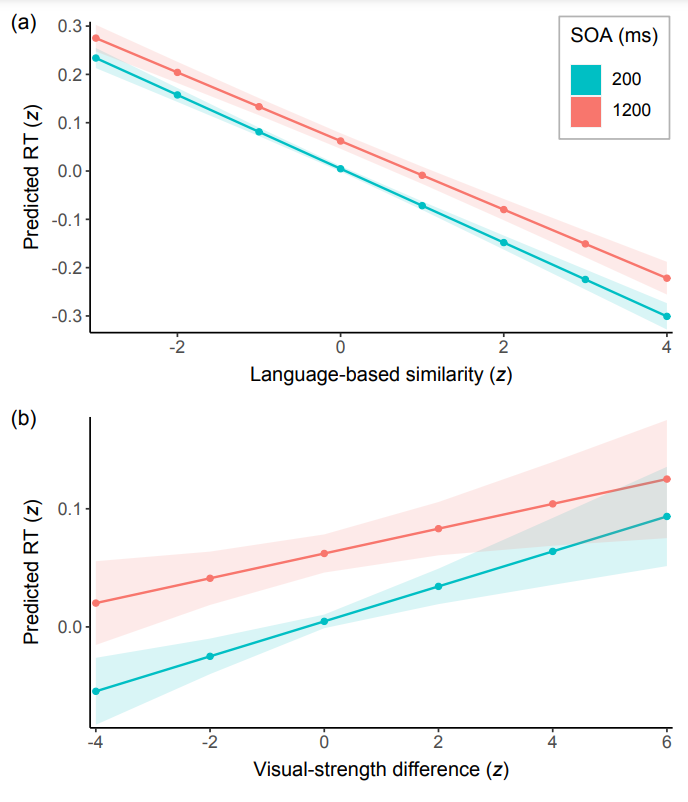
plot_model(fit, type = 'pred', terms = c('spin', 'trait'))
#> Warning: Ignoring unknown parameters: linewidth
...


library(fuzzyjoin)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(knitr)
small_tab = data.frame(Food.Name = c('Corn', 'Squash', 'Peppers'),
Food.Code = c(NA, NA, NA))
large_tab = data.frame(Food.Name = c('Sweet Corn', 'Red Corn', 'Baby Corns',
'Squash', 'Long Squash', 'Red Pepper',
'Green Pepper', 'Red Peppers'),
Food.Code = c(532, 532, 944, 111, 123, 654, 655, 654))
joined_tab = stringdist_join(small_tab, large_tab, by = 'Food.Name',
ignore_case = TRUE, method = 'cosine',
max_dist = 0.5, distance_col = 'dist') %>%
# Tidy columns
select(Food.Name = Food.Name.x, -Food.Name.y,
Food.Code = Food.Code.y, -dist) %>%
# Only keep most frequent food code per food name
group_by(Food.Name) %>% count(Food.Name, Food.Code) %>%
slice(which.max(n)) %>% select(-n) %>%
# Order food names as in the small table
arrange(factor(Food.Name, levels = small_tab$Food.Name))
# Show table with columns renamed
joined_tab %>%
rename('Food Name' = Food.Name,
'Food Code' = Food.Code) %>%
kable()
Food Name
Food Code
Corn
532
Squash
111
Peppers
654
Created on 2023-05-31 with reprex v2.0.2 [Read more...]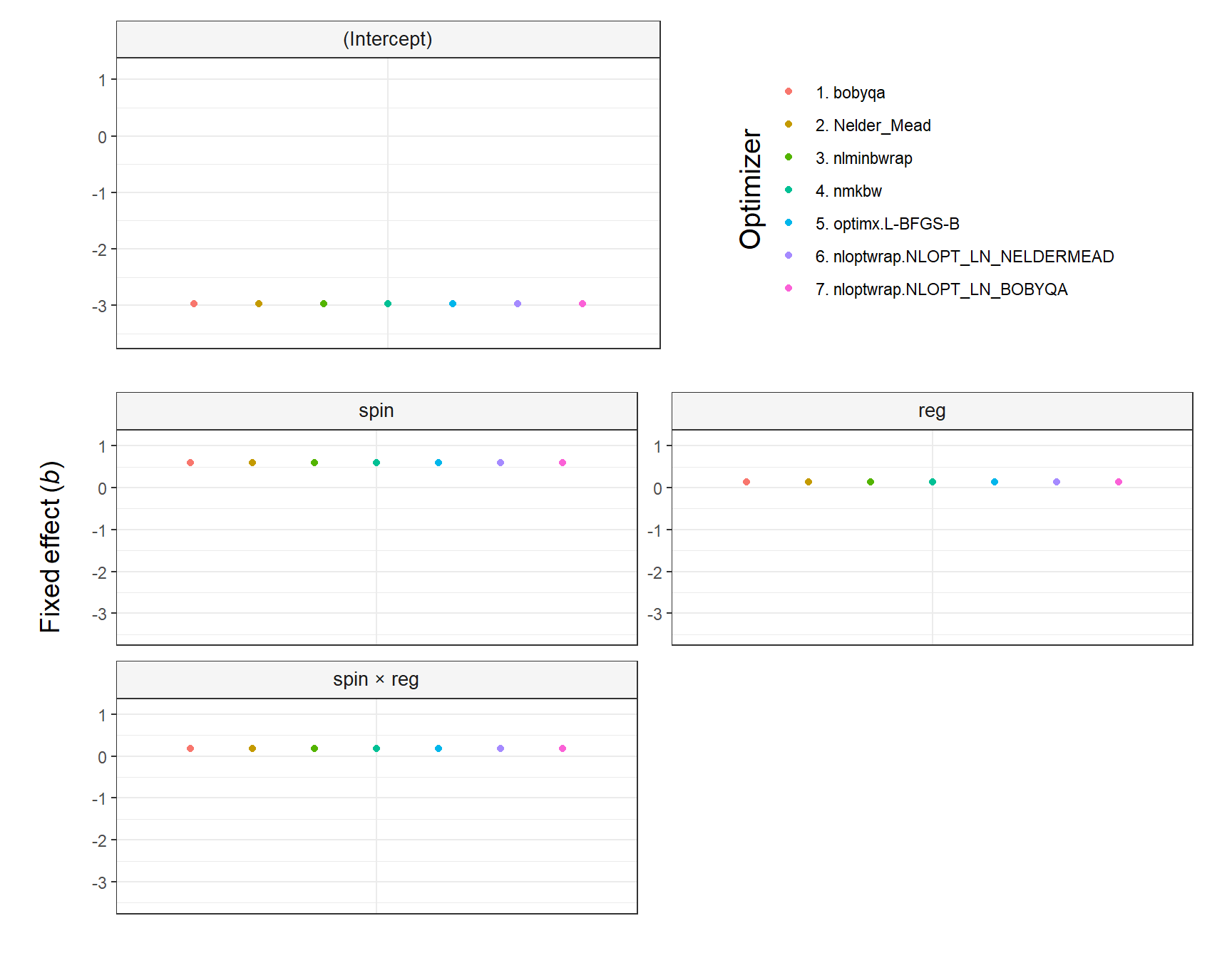
Copyright © 2025 | MH Corporate basic by MH Themes