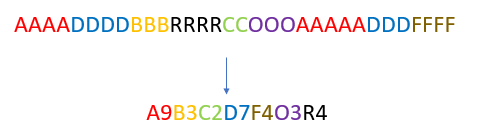Little useless-useful R functions – benchmarking vectors and data.frames on simple GroupBy problem
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
After an interesting conversation on using data. frames or strings (as a vector), I have decided to put this to a simple benchmark test.
The problem is straightforward and simple: a MapReduce or Count with GroupBy.
Problem
Given a simple string of letters:
BBAARRRDDDAAAA
Return a sorted string (per letter) as a key-value pair, with counts per each letter, as:
6A2B3D2R
And in the conversation was the question of optimization of the input data. As the one in a form of string or as an array of rows of data. Sure, anything that is already chunked would perform better. But when?
Data
Let’s create a string of letters and data. frame with letters. Creating the various lengths and consisting of 20 letters (unique).
len <- 100000 # 10^6 lett <- c(LETTERS[1:20]) set.seed(2908) a_vec <- do.call(paste0, c(as.list(sample(lett,len,replace=TRUE)), sep="")) # a vector a_df <- data.frame(chr = paste0(sample(lett,len,replace=TRUE), sep="")) # a data frame
Test
With the rbenchmark framework, I used 20 replications for each test. There are many more solutions to this, initially, I wanted to test the break-point, where string becomes slower than data. frame.
rbenchmark::benchmark(
"table with vector" = {
res_table <- ""
a_table <- table(strsplit(a_vec, ""))
for (i in 1:length(names(a_table))) {
key<- (names(a_table[i]))
val<-(a_table[i])
res_table <- paste0(res_table,key,val)
}
},
"dplyr with data.frame" = {
res_dplyr <- a_df %>%
count(chr, sort=TRUE) %>%
mutate(res = paste0(chr, n, collapse = "")) %>%
select(-chr, -n) %>%
group_by(res)
res_dplyr[1,]
},
"purrr with data.frame" = {
adf_table <- a_df %>%
map(~count(data.frame(x=.x), x))
res_purrr <- ""
for (i in 1:nrow(adf_table$chr)) {
key<- adf_table$chr[[1]][i]
val<- adf_table$chr[[2]][i]
res_purrr <- paste0(res_purrr,key,val)
}
},
replications = 20, order = "relative"
)
Conclusion
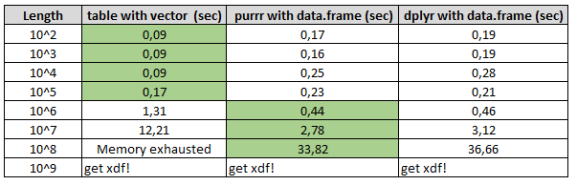
After running 10 tests with different input lengths, the results were on my personal laptop (16GB ram, Windows 10, i5-8th gen, 4 Cores).
Results show that the maximum length that R function strsplit in combination with the table still works fast is for the strings shorter than 1.000.000 characters. Anything larger than this, data. frames will do much better results.
As always, code is available at the Github in the same Useless_R_function repository. Check Github for future updates.
Happy R-coding and stay healthy!“
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.