Learning R: Parameter Fitting for Models Involving Differential Equations
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
It looks like MATLAB, Octave and Python seem to be the preferred tools for scientific and engineering analysis (especially those involving physical models with differential equations). However as part of my learning R experience, I wanted to check out some of R tools for parameter fitting of models involving ordinary differential equations. R has packages deSolve for solving differential equations and FME for parameter fitting. The specific example here is taken from the computational appendix (A.6) of the book Chemical Reactor Analysis and Design Fundamentals by Rawlings and Ekerdt. In fact, all examples in this book are available in Octave and MATLAB.
This example involves two reactions in series
where we need to estimate the rate constants of the the two reactions and
from data
Here the required libraries are loaded
# set working directory
setwd("~/R/wkspace")
# load libraries
library(ggplot2) #library for plotting
library(reshape2) # library for reshaping data (tall-narrow <-> short-wide)
library(deSolve) # library for solving differential equations
library(minpack.lm) # library for least squares fit using levenberg-marquart algorithm
The data available is concentration of A, B, and C over time and is loaded below and plotted
#load concentration data
df=read.table("ABC_data.dat")
names(df)=c("time","ca","cb","cc")
# plot data
tmp=melt(df,id.vars=c("time"),variable.name="species",value.name="conc")
ggplot(data=tmp,aes(x=time,y=conc,color=species))+geom_point(size=3)
The system being solved is
with inital concentrations being .
The rate equations are captured in a function that is an input parameter to the ODE solver
# rate function
rxnrate=function(t,c,parms){
# rate constant passed through a list called parms
k1=parms$k1
k2=parms$k2
# c is the concentration of species
# derivatives dc/dt are computed below
r=rep(0,length(c))
r[1]=-k1*c["A"] #dcA/dt
r[2]=k1*c["A"]-k2*c["B"] #dcB/dt
r[3]=k2*c["B"] #dcC/dt
# the computed derivatives are returned as a list
# order of derivatives needs to be the same as the order of species in c
return(list(r))
}
Computing the predicted concentration for a given set of rate constants involves just solving the ODEs with intial conditions. This is illustrated for the parameter values (which is the actual parameters based on which data was generated).
# predicted concentration for a given parameter set
cinit=c(A=1,B=0,C=0)
t=df$time
parms=list(k1=2,k2=1)
out=ode(y=cinit,times=t,func=rxnrate,parms=parms)
head(out)
time A B C
[1,] 0.000 1.00000000 0.0000000 0.00000000
[2,] 0.263 0.59096447 0.3555550 0.05348051
[3,] 0.526 0.34923934 0.4834497 0.16731099
[4,] 0.789 0.20638798 0.4958219 0.29779013
[5,] 1.053 0.12172441 0.4543303 0.42394528
[6,] 1.316 0.07193491 0.3925423 0.53552284
This is wrapped into a function whose input is the parameters to be estimated and the output is the residuals. Here there are three concentrations that are fitted. In general, we may want to put different weights to these but in this example, they have the same weight. Also, the package FME has several nice features for fitting and uncertainty estimation. Here I have not used FME but directly tried to do the fit more to learn how this works.
ssq=function(parms){
# inital concentration
cinit=c(A=1,B=0,C=0)
# time points for which conc is reported
# include the points where data is available
t=c(seq(0,5,0.1),df$time)
t=sort(unique(t))
# parameters from the parameter estimation routine
k1=parms[1]
k2=parms[2]
# solve ODE for a given set of parameters
out=ode(y=cinit,times=t,func=rxnrate,parms=list(k1=k1,k2=k2))
# Filter data that contains time points where data is available
outdf=data.frame(out)
outdf=outdf[outdf$time %in% df$time,]
# Evaluate predicted vs experimental residual
preddf=melt(outdf,id.var="time",variable.name="species",value.name="conc")
expdf=melt(df,id.var="time",variable.name="species",value.name="conc")
ssqres=preddf$conc-expdf$conc
# return predicted vs experimental residual
return(ssqres)
}
The parameter fitting is done using levenberg-marquardt routine in package minpack.lm.
# parameter fitting using levenberg marquart algorithm # initial guess for parameters parms=c(k1=0.5,k2=0.5) # fitting fitval=nls.lm(par=parms,fn=ssq)
The fitval object has information on estimated parameter and variance covariance matrix
# Summary of fit
summary(fitval)
Parameters:
Estimate Std. Error t value Pr(>|t|)
k1 2.01906 0.04867 41.49 <2e-16 ***
k2 0.99297 0.01779 55.82 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.0212 on 58 degrees of freedom
Number of iterations to termination: 7
Reason for termination: Relative error in the sum of squares is at most `ftol'.
# Estimated parameter
parest=as.list(coef(fitval))
parest
$k1
[1] 2.019065
$k2
[1] 0.992973
# degrees of freedom: # data points - # parameters
dof=3*nrow(df)-2
dof
[1] 58
# mean error
ms=sqrt(deviance(fitval)/dof)
ms
[1] 0.02119577
# variance Covariance Matrix
S=vcov(fitval)
S
k1 k2
k1 0.0023685244 -0.0003605831
k2 -0.0003605831 0.0003164724
The estimated parameters are which is close to the real parameters
used to generate the data. The predicted profiles is overlaid with experimental data
# plot of predicted vs experimental data
# simulated predicted profile at estimated parameter values
cinit=c(A=1,B=0,C=0)
t=seq(0,5,0.2)
parms=as.list(parest)
out=ode(y=cinit,times=t,func=rxnrate,parms=parms)
outdf=data.frame(out)
names(outdf)=c("time","ca_pred","cb_pred","cc_pred")
# Overlay predicted profile with experimental data
tmppred=melt(outdf,id.var=c("time"),variable.name="species",value.name="conc")
tmpexp=melt(df,id.var=c("time"),variable.name="species",value.name="conc")
p=ggplot(data=tmppred,aes(x=time,y=conc,color=species,linetype=species))+geom_line()
p=p+geom_line(data=tmpexp,aes(x=time,y=conc,color=species,linetype=species))
p=p+geom_point(data=tmpexp,aes(x=time,y=conc,color=species))
p=p+scale_linetype_manual(values=c(0,1,0,1,0,1))
p=p+scale_color_manual(values=rep(c("red","blue","green"),each=2))+theme_bw()
print(p)
Estimation of Parameter Uncertainty
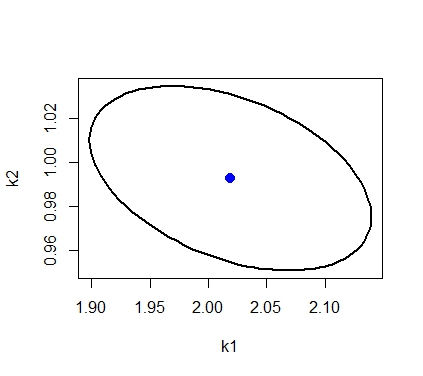
Usually though the model is nonlinear, the confidence region is approximated using the following ellipsoid
where is the estimated value of parameter,
is the variance covariance matrix of estimated parameters and
is the number of parameters and
is the total number of data points.
# draw the confidence region # get points for a circle with radius r r=sqrt(qf(0.95,2,58)*2) theta=seq(0,2*pi,length.out=100) z=cbind(r*cos(theta),r*sin(theta)) # transform points of circle into points of ellipse using # svd of inverse covariance matrix Sinv_svd=svd(Sinv) # inverse of covariance matrix xt=t(Sinv_svd$v)%*%diag(1/sqrt(Sinv_svd$d))%*%t(z) # transform from circle to ellispse x=t(xt) # translate the ellipse so that center is the estimated parameter value x=x+matrix(rep(as.numeric(parest),100),nrow=100,byrow=T) plot(x[,1],x[,2],type="l",xlab="k1",ylab="k2",lwd=2) points(parest$k1,parest$k2,pch=20,col="blue",cex=2)
Another way of estimating the uncertainty is using bootstrapping procedure. Here several simulated datasets are generated using the current estimated model and adding random normal noise to each data point (with mean=0 and variance = mean square error from model). Then the parameter is estimated for each simulated dataset. The set of parameters thus generated indicate the uncertainty in parameter estimates.
# Simulation based estimation of uncertainty
# store original experimental data in a separate dataframe
dforig=df
# conc profile based on estimated k1 and k2
cinit=c(A=1,B=0,C=0)
t=dforig$time
parms=parest
out=ode(y=cinit,times=t,func=rxnrate,parms=parms)
outsim=matrix(0,nrow=nrow(dforig),ncol=4)
outsim[,1]=out[,1]
# number of simulations
nsim=1000
parsim=matrix(0,nrow=nsim,ncol=2)
colnames(parsim)=c("k1","k2")
for (i in 1:nsim){
# Simulate data set by adding normal random variable with mean 0 and stdev from fit
outsim[,2:4]=out[,2:4]+matrix(rnorm(3*nrow(dforig)),nrow=nrow(dforig),ncol=3)*ms
df=data.frame(outsim)
names(df)=c("time","ca","cb","cc")
# get parameter estimate for the simulated dataset
parms=as.numeric(parest)
fitsim=nls.lm(par=parms,fn=ssq)
# store estimated parameters in the ith row
parsim[i,]=coef(fitsim)
}
# plot the parameter estimates from the 1000 simulations
plot(parsim[,1],parsim[,2],xlab="k1",ylab="k2")
# overlay the 95% ellipse computed previously
lines(x[,1],x[,2],col="blue",lwd=2)
Next, the percentage of parameter estimates that fall within the ellipse is computed and found to be 93% (expected is 95%). In this case, the ellipsoidal approximation of parameter uncertainty seems adequate. But this might not be the case in general.
# percentage of parameters from simulation within the 95% ellipse
tmp=rep(0,length.out=nsim)
for(i in 1:nsim){
tmp[i]=(parsim[i,]-as.numeric(parest))%*%Sinv%*%(parsim[i,]-as.numeric(parest))
}
sum(tmp <= qf(0.95,2,58)*2)/nsim
[1] 0.933
As I mentioned previously, the package FME has several functions for parameter fitting, MCMC simulation, sensitivity analysis but was not used in this post. Also, when running multiple simulations using ODE models, it might be better to use the C compiled version of reaction rate function. deSolve package provides that ability and describes it in this vignette. The data and R code is available here.
# session Info sessionInfo() R version 3.0.1 (2013-05-16) Platform: i386-w64-mingw32/i386 (32-bit) locale: [1] LC_COLLATE=English_United States.1252 LC_CTYPE=English_United States.1252 [3] LC_MONETARY=English_United States.1252 LC_NUMERIC=C [5] LC_TIME=English_United States.1252 attached base packages: [1] stats graphics grDevices utils datasets methods base other attached packages: [1] minpack.lm_1.1-7 deSolve_1.10-6 reshape2_1.2.2 ggplot2_0.9.3.1 loaded via a namespace (and not attached): [1] colorspace_1.2-2 dichromat_2.0-0 digest_0.6.3 grid_3.0.1 gtable_0.1.2 [6] labeling_0.2 MASS_7.3-26 munsell_0.4 plyr_1.8 proto_0.3-10 [11] RColorBrewer_1.0-5 scales_0.2.3 stringr_0.6.2 tools_3.0.1
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.