Tutorial- Building Biological Networks
[This article was first published on imDEV » R, and kindly contributed to R-bloggers]. (You can report issue about the content on this page here)
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
I love networks! Nothing is better for visualizing complex multivariate relationships be it social, virtual or biological.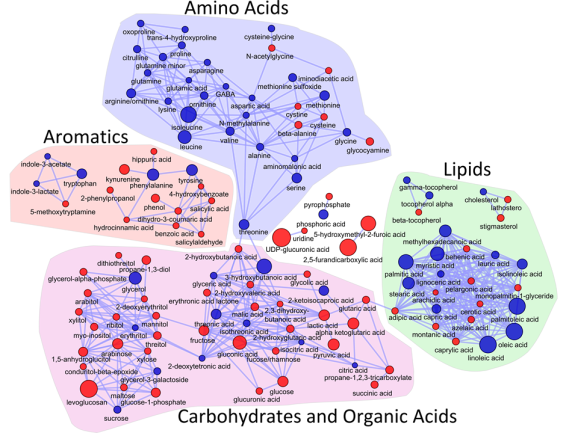
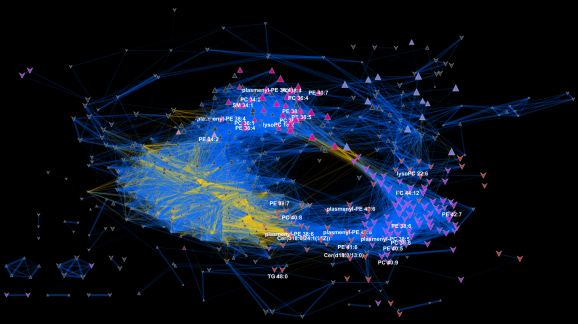
I recently gave a hands-on network building tutorial using R and Cytoscape to build large biological networks. In these networks Nodes represent metabolites and edges can be many things, but I specifically focused on biochemical relationships and chemical similarities. Your imagination is the limit.
If you are interested check out the presentation below.
Here is all the R code and links to relevant data you will need to let you follow along with the tutorial.
#load needed functions: R package in progress – “devium”, which is stored on github source(“http://pastebin.com/raw.php?i=Y0YYEBia”)# get sample chemical identifiers here:https://docs.google.com/spreadsheet/ccc?key=0Ap1AEMfo-fh9dFZSSm5WSHlqMC1QdkNMWFZCeWdVbEE#gid=1 #Pubchem CIDs = cids cids # overview nrow(cids) # how many str(cids) # structure, wan't numeric cids<-as.numeric(as.character(unlist(cids))) # hack to break factor #get KEGG RPAIRS #making an edge list based on CIDs from KEGG reactant pairs KEGG.edge.list<-CID.to.KEGG.pairs(cid=cids,database=get.KEGG.pairs(),lookup=get.CID.KEGG.pairs()) head(KEGG.edge.list) dim(KEGG.edge.list) # a two column list with CID to CID connections based on KEGG RPAIS # how did I get this? #1) convert from CID to KEGG using get.CID.KEGG.pairs(), which is a table stored:https://gist.github.com/dgrapov/4964546 #2) get KEGG RPAIRS using get.KEGG.pairs() which is a table stored:https://gist.github.com/dgrapov/4964564 #3) return CID pairs #get EDGES based on chemical similarity (Tanimoto distances >0.07) tanimoto.edges<-CID.to.tanimoto(cids=cids, cut.off = .7, parallel=FALSE) head(tanimoto.edges) # how did I get this? #1) Use R package ChemmineR to querry Pubchem PUG to get molecular fingerprints #2) calculate simialrity coefficient #3) return edges with similarity above cut.off #after a little bit of formatting make combined KEGG + tanimoto edge list # https://docs.google.com/spreadsheet/ccc?key=0Ap1AEMfo-fh9dFZSSm5WSHlqMC1QdkNMWFZCeWdVbEE#gid=2 #now upload this and a sample node attribute table (https://docs.google.com/spreadsheet/ccc?key=0Ap1AEMfo-fh9dFZSSm5WSHlqMC1QdkNMWFZCeWdVbEE#gid=1) #to Cytoscape
You can also download all the necessary materials HERE, which include:
- tutorial in powerpoint
- R script
- Network edge list and node attributes table
- Cytoscape file
Happy network making!
To leave a comment for the author, please follow the link and comment on their blog: imDEV » R.
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.