Advent of 2021, Day 19 – Data engineering for Spark Streaming
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
Series of Apache Spark posts:
- Dec 01: What is Apache Spark
- Dec 02: Installing Apache Spark
- Dec 03: Getting around CLI and WEB UI in Apache Spark
- Dec 04: Spark Architecture – Local and cluster mode
- Dec 05: Setting up Spark Cluster
- Dec 06: Setting up IDE
- Dec 07: Starting Spark with R and Python
- Dec 08: Creating RDD files
- Dec 09: RDD Operations
- Dec 10: Working with data frames
- Dec 11: Working with packages and spark DataFrames
- Dec 12: Spark SQL
- Dec 13: Spark SQL Bucketing and partitioning
- Dec 14: Spark SQL query hints and executions
- Dec 15: Introduction to Spark Streaming
- Dec 16: Dataframe operations for Spark streaming
- Dec 17: Watermarking and joins for Spark streaming
- Dec 18: Time windows for Spark streaming
Streaming data can be used in conjunction with other datasets. You can have Joining streaming data, joining data with watermarking, deduplication, outputting the data, applying foreach logic, using triggers and creating Stream API Tables.
All of the functions are available in Python, Scala and Java and some are not available with R. We will be focusing on Python and R.
Joining data
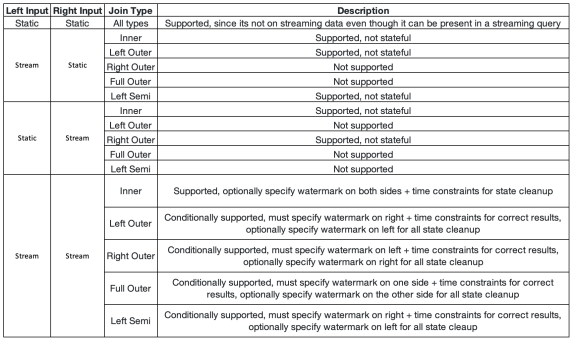
Structured Streaming supports joining a streaming dataframe with either a static dataFrame or another streaming dataframe (or both, or multiple).There are inner, outer, and semi joins (and some others) and the result of a join with streaming or static dataframe will have the static or streaming data.
With stream join in Python (pseudo code), you can simply do:
staticDf = spark.read. ... streamingDf = spark.readStream. ... streamingDf.join(staticDf, "type") # inner equi-join with a static DF streamingDf.join(staticDf, "type", "left_outer") # left outer join with a static DF
or with using R:
staticDf <- read.df(...)
streamingDf <- read.stream(...)
joined <- merge(streamingDf, staticDf, sort = FALSE) # inner equi-join with a static DF
joined <- join(
streamingDf,
staticDf,
streamingDf$value == staticDf$value,
"left_outer") # left outer join with a static DF
You can also use inner joins with watermarking, but there are some cautions. When the record is growing indefinitely and all the data must be saved, it can happen that the unbounded state appear. Therefore you have to define additional join condition what to do with old data that cannot match incoming data and how to clean the state. So you need to define watermark delay on both inputs and define a constraint on event-time across the two inputs, so that the engine can figure out when the old rows will not be required to match with the new (or upcoming) rows.
In Python (as well as in R) you have to define the watermarking delay and event-time range condition.
from pyspark.sql.functions import expr
impressions = spark.readStream. ...
clicks = spark.readStream. ...
# Apply watermarks on event-time columns
impressionsWithWatermark = impressions.withWatermark("impressionTime", "2 hours")
clicksWithWatermark = clicks.withWatermark("clickTime", "3 hours")
# Join with event-time constraints
impressionsWithWatermark.join(
clicksWithWatermark,
expr("""
clickAdId = impressionAdId AND
clickTime >= impressionTime AND
clickTime <= impressionTime + interval 1 hour
""")
)
Same rules apply to R:
impressions <- read.stream(...)
clicks <- read.stream(...)
# Apply watermarks on event-time columns
impressionsWithWatermark <- withWatermark(impressions, "impressionTime", "2 hours")
clicksWithWatermark <- withWatermark(clicks, "clickTime", "3 hours")
# Join with event-time constraints
joined <- join(
impressionsWithWatermark,
clicksWithWatermark,
expr(
paste(
"clickAdId = impressionAdId AND",
"clickTime >= impressionTime AND",
"clickTime <= impressionTime + interval 1 hour"
)))
When using the outer join, you have to define watermarking and event-time constraints, mainly because of the NULL values. When using semi-joins, also the watermarking and event-time constraints must be defined. So only in case of inner join, there is no need to define watermarking and event-time constraint.
Here is the matrix for semi-joins:
Deduplication
To deduplicate the streaming records, best way to do it is by using a unique identifies in the events. Same as on static dataframes and using a unique identifier column. Deduplication can be used with or without watermarking. With watermarking you define the boundaries on how late a duplicate record may arrive and then you define a watermark on event-time column and deduplicate both the unique identifier and time column. If you are not using watermark, this means there are no boundaries on when to deduplicate or on when a duplicate may be ingested and therefore the query will hold all the past records (until further purged)
Let’s check the deduplication with Python (using pseudo code and using guid as unique identifier):
streamingDf = spark.readStream. ...
# Without watermark using guid column
streamingDf.dropDuplicates("guid")
# With watermark using guid and eventTime columns
streamingDf \
.withWatermark("eventTime", "10 seconds") \
.dropDuplicates("guid", "eventTime")
And with R:
streamingDf <- read.stream(...) # Without watermark using guid column streamingDf <- dropDuplicates(streamingDf, "guid") # With watermark using guid and eventTime columns streamingDf <- withWatermark(streamingDf, "eventTime", "10 seconds") streamingDf <- dropDuplicates(streamingDf, "guid", "eventTime")
Outputting the data
State store is a versioned key-value store which provides both read and write operations. In Structured Streaming, we use the state store provider to handle the stateful operations across batches. When outputing the streaming queries there are three modes: append or default mode, complete mode, and update mode.
Append mode will only add the new rows to the result table since the last trigger was outputted to the sink. This mode is supported only to type of data and the rows, where there will never be any change. Because this mode guaranties that each row will be in the output only once. Queries using select, where, map, flatMap, filter, join (and others) will support append mode.
Complete mode will make the whole result table outputted every time the trigger is finished. This is supported for aggregated queries, because the results will be different.
Update mode will have only the rows that were updated since the last trigger was executed will be outputted to the sink from the result table.
Output sinks can be: file sink, Kafka sink, console and memory sink (both for debugging purposes) and foreach sink.
So let’s assume with pseudo Python code that we are not doing any aggregation and we are storing data to console sink or file sink
# ========== DF with no aggregations ==========
noAggDF = deviceDataDf.select("device").where("signal > 10")
# Print new data to console
noAggDF \
.writeStream \
.format("console") \
.start()
# Write new data to Parquet files
noAggDF \
.writeStream \
.format("parquet") \
.option("checkpointLocation", "path/to/checkpoint/dir") \
.option("path", "path/to/destination/dir") \
.start()
If there were aggregations, different output mode and sinks must be chosen. And for R example, let’s take the aggregations and we are using both complete output mode (remember the aggregations) and using in-memory or console sink.
# ========== DF with aggregation ==========
aggDF <- count(groupBy(df, "device"))
# Print updated aggregations to console
write.stream(aggDF, "console", outputMode = "complete")
# Have all the aggregates in an in memory table. The query name will be the table name
write.stream(aggDF, "memory", queryName = "aggregates", outputMode = "complete")
# Interactively query in-memory table
head(sql("select * from aggregates"))
Foreach and ForeachBatch
Foreach or ForeachBatch operations will give you apply operator to your streaming query. So for every row, there can be a logic, calculation, aggregation applied. When using Foreach operator this applies for every row, with ForeachBatch, it will be applied on each batch.
Let’s look into Foreach with Python (using pseudo code):
def process_row(row):
# Write or apply a logic to a row
pass
query = streamingDF.writeStream.foreach(process_row).start()
In un-similar fashion, this operator can not be applied to R. 
Triggers
The trigger settings of a streaming query define the timing of streaming data processing. Trigger define how the query is going to be executed. And since it is a time bound, it can execute a query as batch query with fixed interval or as a continuous processing query.
Spark Streaming gives you three types of triggers: Fixed interval micro-batches, one time micro-batch, and continuous with fixed intervals.
Using Python, here are all three type of triggers (again as a pseudo code):
# Default trigger (runs micro-batch as soon as it can)
df.writeStream \
.format("console") \
.start()
# ProcessingTime trigger with two-seconds micro-batch interval
df.writeStream \
.format("console") \
.trigger(processingTime='2 seconds') \
.start()
# One-time trigger
df.writeStream \
.format("console") \
.trigger(once=True) \
.start()
# Continuous trigger with one-second checkpointing interval
df.writeStream
.format("console")
.trigger(continuous='1 second')
.start()
Two type of triggers are also available in R. The micro-batch and one-time triggers. Continous trigger is not (yet) supported with R. Here is the pseudo code:
# Default trigger (runs micro-batch as soon as it can) write.stream(df, "console") # ProcessingTime trigger with two-seconds micro-batch interval write.stream(df, "console", trigger.processingTime = "2 seconds") # One-time trigger write.stream(df, "console", trigger.once = TRUE)
There are some even more details on data engineering and different operators, but this post was created to cover the most important ones.
Tomorrow we will look into Spark graph processing.
Compete set of code, documents, notebooks, and all of the materials will be available at the Github repository: https://github.com/tomaztk/Spark-for-data-engineers
Happy Spark Advent of 2021! 
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.