Simulating metapopulation occupation in a landscape
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
The objective of this post is to go into the inner workings of the package MetaLandSim, which I developed a few years ago. MetaLandSim’s main objectives are to i) simulate the occupation of an habitat network suffering some sort of change (but static landscapes work too); ii) simulate range expansion by a species with a metapopulation-like spatial strategy. Why the emphasis on metapopulation? Well, because these “landscapes” are, in reality graph-like simplifications of a landscape (if you don’t know what a graph is, check here). You can also check the package manual for details.
MetaLandSim simulates the stochastic occupancy of the landscape by a given species using Stochastic Patch Occupancy Models, like the Incidence Function Model, developed by Hanski (1994). Being a simulation based upon a stochastic model, it produces slightly different results with each simulation. That’s why we have to repeat each parameter combination many times. Here, considering this is a demonstration, I will run only one simulation.
First, as always, load the package:
library(MetaLandSim)
Then we should define the characteristics of the graph-landscape. Here we are creating a landscape square with a size of 5000×5000 meters, minimum distance between patches of 20 meters, mean area 0.8 hectares and we are using 300 -which I called dispersal ability- as a threshold to aggregate habitat patches).
Running this function (if plotG=TRUE) will immediately plot the landscape-graph:
rl1 <- rland.graph(mapsize=5000, dist_m=20, areaM=0.8,
areaSD=0.2, Npatch=180,
disp=300, plotG=TRUE)

There’s no need to re-plot it now, considering that the function rland.graph already did it, but if (for some reason) that’s necessary, here’s how:
plot_graph(rl1, species=FALSE, links=TRUE)
And now, we can create the first occupied landscape (here I selected 10% of the patches to be occupied, those patches in green):
sp1 <- species.graph(rl=rl1, method="number", parm=10, nsew="none", plotG=TRUE)
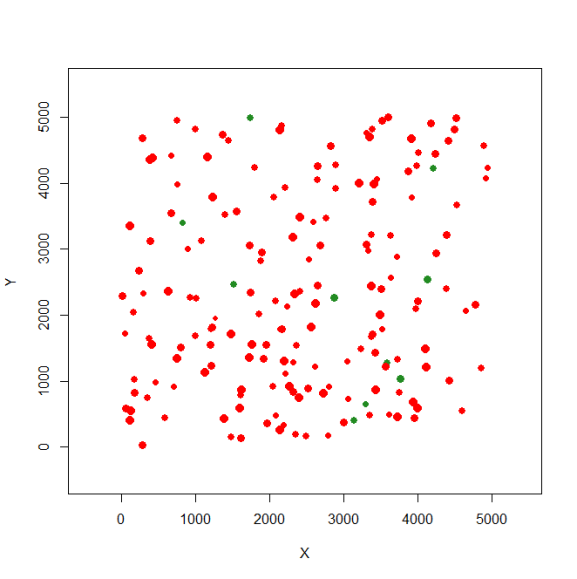
Again, now we don’t need to re-plot this landscape. But if that’s necessary use this:
plot_graph(sp1, species=TRUE, links=FALSE)
Starting with the previously created landscape (rl1) we now create a list containing this landscape through 100 time steps (here I did not consider any dynamics -par1=”none”-, check the manual for details, so all time steps have the same landscape):
span1 <- span.graph(rl=rl1, span=100, par1="none", par2=NULL, par3=NULL, par4=NULL, par5=NULL)
Now we need to define the species parameters, defining its metapopulational dynamics.
Metalandsim provides Bayesian-based functions to derive these parameters, based upon the work of Benjamin Risk in this paper (his original functions are here).
Benjamin adapted his functions to be included in this package. Later, we collaborated, and used this approach in our 2017 paper (we provided the R scripts as supplementary information, these can be downloaded from the paper’s website). This is the recommended procedure, but the package has another function (parameter.estimate). It provides some other approaches (however I don’t recommend it). It is useful sometimes, for didactic reasons, because it’s way simpler (and that is why it was not yet deprecated).
Here I’m only going to create the parameter data frame, assuming these were previously estimated.
param1 <- create.parameter.df(alpha=0.0045, x=0.5, y=2, e=0.04)
For details on what does each of these parameters means go here.
And, finally, simulate the species occupation (considering the parameters just defined) through the time steps:
sim1 <- simulate_graph(rl=sp1, rlist=span1, simulate.start=FALSE, method=NULL, parm=NULL, nsew="none", succ = "none", param_df=param1, kern="op1", conn="op1", colnz="op1", ext="op1", beta1=NULL, b=1, c1=NULL, c2=NULL, z=NULL, R=NULL )
No, just to see what with the species occupation thorough time, we can plot a few of the time steps (1, 20, 40, 60, 80 and 100):
par(mfrow=c(2,3))#we are going to 6 time steps #First landscape plotL.graph(rl=sp1, rlist=sim1, #time step 1 nr=1, species=TRUE, links=FALSE) plotL.graph(rl=sp1, rlist=sim1, #time step 20 nr=20, species=TRUE, links=FALSE) plotL.graph(rl=sp1, rlist=sim1, #time step 40 nr=40, species=TRUE, links=FALSE) plotL.graph(rl=sp1, rlist=sim1, #time step 60 nr=60, species=TRUE, links=FALSE) plotL.graph(rl=sp1, rlist=sim1, #time step 80 nr=80, species=TRUE, links=FALSE) plotL.graph(rl=sp1, rlist=sim1, #time step 100 nr=100, species=TRUE, links=FALSE)
The species gradually expands from the initial occupied habitat patches (the green patches on the top left landscape-graph):

Finally, just a few reminders:
- This post was only to demonstrate the basic reasoning behind these simulations;
- The species parameters should be carefully estimated (see the Bayesian approach);
- MetaLandSim’s simulations should be repeated many times (to allow the results to stabilize). This one-run simulation was an example (for more on this, check the iterate.graph function).
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
