Using and interpreting different contrasts in linear models in R
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
When building a regression model with categorical variables with more than two levels (ie “Cold”, “Freezing”, “Warm”) R is doing internally some transformation to be able to compute regression coefficient. What R is doing is that it is turning your categorical variables into a set of contrasts, this number of contrasts is the number of level in the variable (3 in the example above) minus 1. Here I will present three ways to set the contrasts and depending on your research question and your variables one might be more appropriate than the others.
# setting seed so that numerical results stay stable
set.seed(25)
# let's imagine an experiment which measure plant biomass based on various
# levels of nutrient added to the medium first case one treatments three
# levels
f <- gl(n = 3, k = 20, labels = c("control", "low", "high"))
# with treatments contrasts (default)
mat <- model.matrix(~f, data = data.frame(f = f))
# this tell us which contrast has been used to generate the model matrix
attr(mat, "contrasts")
## $f
## [1] "contr.treatment"
# simulate some ys
beta y m |t|)
## (Intercept) 11.498 0.463 24.84 < 2e-16 ***
## flow 3.037 0.655 4.64 2.1e-05 ***
## fhigh 6.163 0.655 9.41 3.3e-13 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 2.07 on 57 degrees of freedom
## Multiple R-squared: 0.609, Adjusted R-squared: 0.595
## F-statistic: 44.3 on 2 and 57 DF, p-value: 2.45e-12
The first coefficient, the intercept, is the estimated average for the baseline group, which in our case is the control group (in the control group the biomass is estimated to be on average 11.49). The second coefficient “flow” is the estimated difference between the average in the baseline group and the average in the “low” group (this group has an increase in 3.03 biomass compared to the control group). Similarly the third coefficient is the difference between the average in the baseline and the “high” group (this group has an increase of 6.16 biomass compared to the control group).
# plot the results plot(y ~ rep(1:3, each = 20), xaxt = "n", xlab = "Treatment") axis(1, at = c(1, 2, 3), labels = levels(f)) points(c(1, 2, 3), c(coef(m)[1], coef(m)[2:3] + coef(m)[1]), pch = 16, cex = 2)
This is by default, now let’s turn to other contrasts options:
# now with helmert contrasts mat attr(mat, "contrasts") ## $f ## [1] "contr.helmert" # simulate the ys beta <- c(5, 3, 2) y <- rnorm(n = 60, mean = mat %*% beta, sd = 1.5) # model m <- lm(y ~ f, contrasts = list(f = "contr.helmert")) #there we tell the model to use helmert contrast to build the model summary(m) ## ## Call: ## lm(formula = y ~ f, contrasts = list(f = "contr.helmert")) ## ## Residuals: ## Min 1Q Median 3Q Max ## -2.281 -0.973 -0.202 0.752 2.986 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 4.756 0.184 25.9 <2e-16 *** ## f1 3.116 0.225 13.9 <2e-16 *** ## f2 1.949 0.130 15.0 <2e-16 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 1.42 on 57 degrees of freedom ## Multiple R-squared: 0.88, Adjusted R-squared: 0.876 ## F-statistic: 209 on 2 and 57 DF, p-value: <2e-16
Now the meaning of the various coefficient is different, the intercept is the average biomass over all the levels (across control, low and high), f1 is the difference between the average of the first level (control) and the average of the second one (low), plants in the treatment low have a 3.11 increase in biomass. f2 is the difference between the average of control and low and the average of high treatment. To put this differently, if we put together the data from the control and low treatment and compare there average value to the average value of plants in the high treatment we would get fhigh. Mean biomass of the plants in the high treatment is higher by 1.95 than plants of the control and low treatment. This type of contrast is a bit harder to interpret but is well suited for variables where the levels have an order, ie (“0”,“>0 and <5″,”>5 and <10” …), there we can gradually compare the successive levels.
As a note, in most applied cases we can set the contrasts that will be used by the model using contrasts(factor)<-‘some_contrasts’. Let’s turn to the last contrasts the effect (or deviation) coding:
# now with sum contrasts, let's spice up a little bit and introduce an
# interaction with a continuous variables
x <- runif(60, -2, 2)
levels(f) <- c("North", "South", "Center") #let's make different level which cannot easily be given an order or a baseline
mat <- model.matrix(~x * f, data = data.frame(f = f), contrasts.arg = list(f = "contr.sum"))
attr(mat, "contrasts")
# simulate the ys
beta <- c(5, -2, 3, 2, 1, -4)
y <- rnorm(n = 60, mean = mat %*% beta, sd = 1.5)
# model
m <- lm(y ~ x * f, contrasts = list(f = "contr.sum"))
summary(m)
##
## Call:
## lm(formula = y ~ x * f, contrasts = list(f = "contr.sum"))
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.271 -1.021 -0.165 0.867 3.914
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 5.057 0.207 24.42 < 2e-16 ***
## x -1.869 0.185 -10.13 4.4e-14 ***
## f1 2.699 0.295 9.16 1.4e-12 ***
## f2 2.351 0.293 8.03 8.8e-11 ***
## x:f1 0.954 0.264 3.61 0.00067 ***
## x:f2 -4.269 0.268 -15.92 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.58 on 54 degrees of freedom
## Multiple R-squared: 0.922, Adjusted R-squared: 0.914
## F-statistic: 127 on 5 and 54 DF, p-value: <2e-16
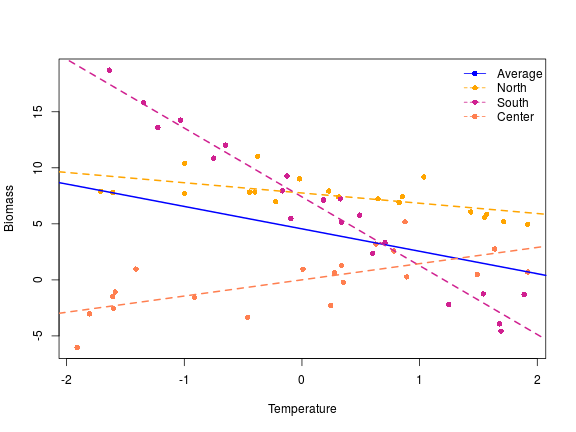
This type of contrasts is useful when there is no natural way to set a baseline or an ordering in the different levels of the variables. I changed the name of the level to illustrate what I mean by this, let’s imagine in this context that we had sampled our plant in three different localities, it is hard to determine in this context what should be the baseline, the deviation coding is a nice way to model these type of data. The intercept in this context is the overall mean across the levels (as in the helmert contrasts), so overall the plant biomass was 5.05. The second one the the average slope between the biomass and the x variable, if we increase x by one the plant biomass decrease by -1.87 across the geographical gradient. f1 is the difference between the overall mean and the mean in the north locality, similarly f2 is the difference between the overall mean and the south locality. To get the estimated average value at the center locality we have to do:
coef(m)[1] - coef(m)[3] - coef(m)[4] ## (Intercept) ## 0.00678
The interaction coefficient are the deviation of the slope within a group from the overall slope, therefore in the north if we increase x by 1, we decrease the biomass by 1.87-0.95= 0.92, similarly the slope in the south is -1.87+(-4.27)= -6.14 and in the center: -1.87-0.95-(-4.27) = +1.45. Around each of these coefficient we have some assessement of the significance of the difference between the overall mean and the various groups. So far I could not find a way to assess the significance of the difference between the overall mean and the last group …
Let’s do a nice figure of this:
# a nice plot
plot(y ~ x, xlab = "Temperature", ylab = "Biomass", pch = 16, col = rep(c("orange",
"violetred", "coral"), each = 20))
abline(a = 4.55, b = -2, lwd = 2, lty = 1, col = "blue")
abline(a = coef(m)[1] + coef(m)[3], b = coef(m)[2] + coef(m)[5], lwd = 2, lty = 2,
col = "orange")
abline(a = coef(m)[1] + coef(m)[4], b = coef(m)[2] + coef(m)[6], lwd = 2, lty = 2,
col = "violetred")
abline(a = coef(m)[1] - coef(m)[3] - coef(m)[4], b = coef(m)[2] - coef(m)[5] -
coef(m)[6], lwd = 2, lty = 2, col = "coral")
legend("topright", legend = c("Average", "North", "South", "Center"), col = c("blue",
"orange", "violetred", "coral"), pch = 16, lty = c(1, 2, 2, 2), bty = "n")
Thats’s it for today, this page was greatly helpful in understanding the meaning of the various contrasts: http://www.unc.edu/courses/2006spring/ecol/145/001/docs/lectures/lecture26.htm, the wikipedia page is also rather nice on this: https://en.wikipedia.org/wiki/Categorical_variable
Filed under: R and Stat Tagged: contrasts, LM, R
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.